R.utils::sourceDirectory(here::here("src", "functions"))Multi-panel plots: Faceting
Overview
When you map variables to aesthetics, you get legends. Legends are certainly helpful but legends cannot remove the plot clutter associated with many subgroups of data corresponding to those levels in the legend. Sometimes, you want to plot individual plots for various subgroups (e.g., each state, student, year, etc.) rather than map a variable containing those subgroups to a plot aesthetic. This is where we create, small multiples, or small versions of the same plot for each subgroup which are then combined into a grid arrangement (e.g., rows, columns). The advantage is that the plots are easier to process. A disadvantage comes with making comparisons across the individual plots when they are not on aligned scales. For example, comparing data on the y-axis is fairly easy to do when plots are arranged in a row (e.g., compare the height position of bars) but when plots are arranged in a column, this comparison is complicated. Heights of bars cannot be compared but instead the length of the bars need to be extracted for comparison. When plots of interest are arranged diagonally, comparing data on either x or y axis is more demanding. This cognitive process is more demanding, thus errors of interpretation can be made. This outcome is a consequence of plots composed small multiples. Nevertheless, lots of individual data can be presented using small multiples and general pattern matching or mismatching processes can be performed quite easily.
To Do
Review corresponding canvas content.
Readings
External Functions
Provided in class:
view_html(): for viewing data frames in html format, from /src/my_functions.R
You can use this in your own work space but I am having a challenge rendering this of the website, so I’ll default to print() on occasion.
Libraries
- {dplyr} 1.1.4: for selecting, filtering, and mutating
- {forcats} 1.0.0: for creating and ordering factors
- {ggplot2} 3.5.1: for plotting
Loading Libraries
pacman::p_load(tidyverse)Loading Data
To examine some associations, we will use hammer throwing distances times which can be accessed from:
To access the data, either read the file directly from the url using read.csv() and assign the data frame a name like DATA:
read.csv("https://github.com/slicesofdata/dataviz24/raw/main/data/tfrrs/hammer_data.csv")
Small Multiples, Faceting, and Multiple Panels
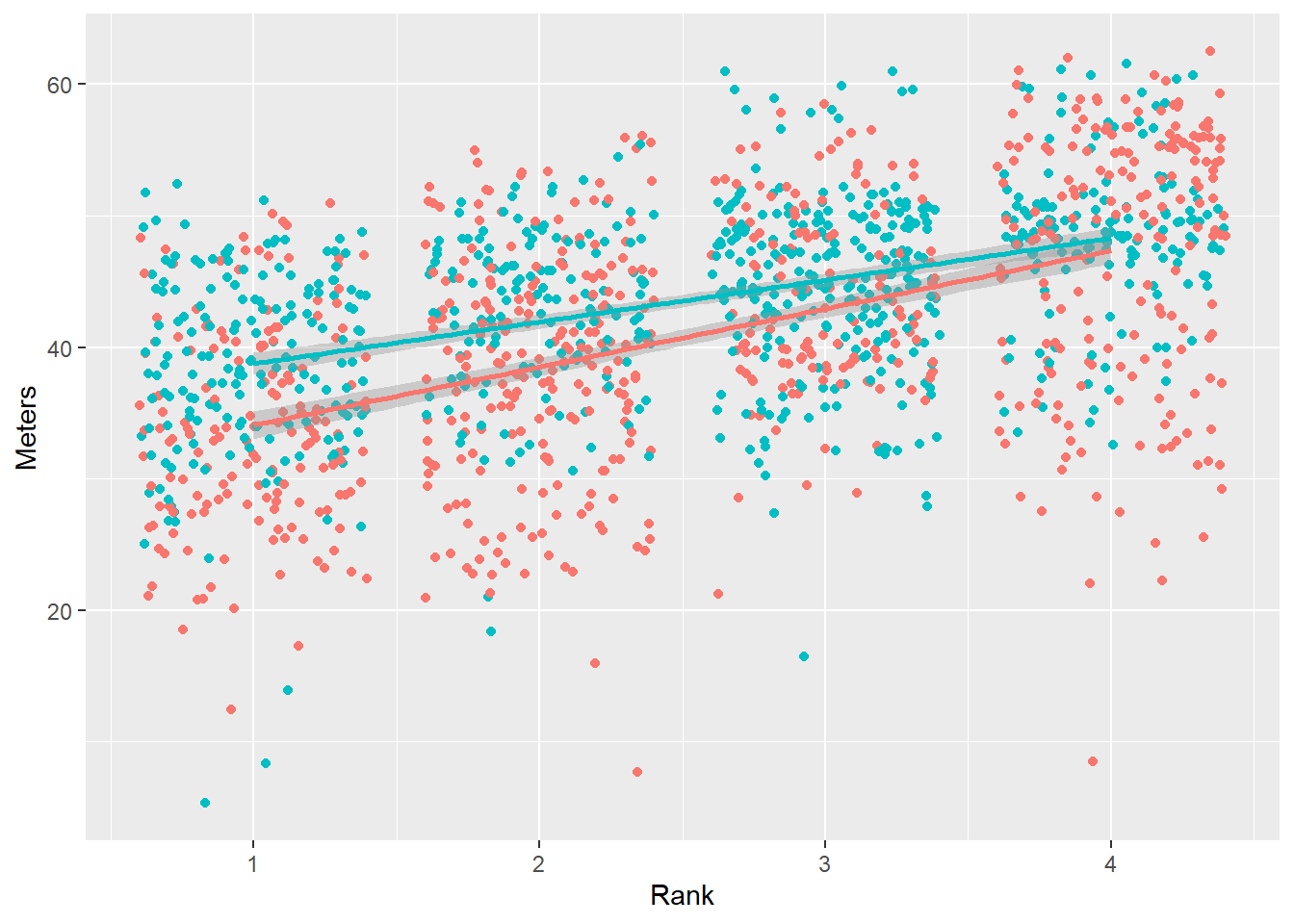
We will create a base plot for passing to facet layers. The linear is used for illustration purposes. You must know when applying a linear model is appropriate given your data.
(base_plot <-
DATA |>
filter(!is.na(Rank)) |>
ggplot(mapping = aes(x = Rank,
y = Meters,
color = Team
)) +
geom_point(position = position_jitter()) +
geom_smooth(mapping = aes(color = Team),
method = "lm",
fullrange = TRUE
) +
theme(legend.position = "none")
)
Creating facets using facet_wrap()
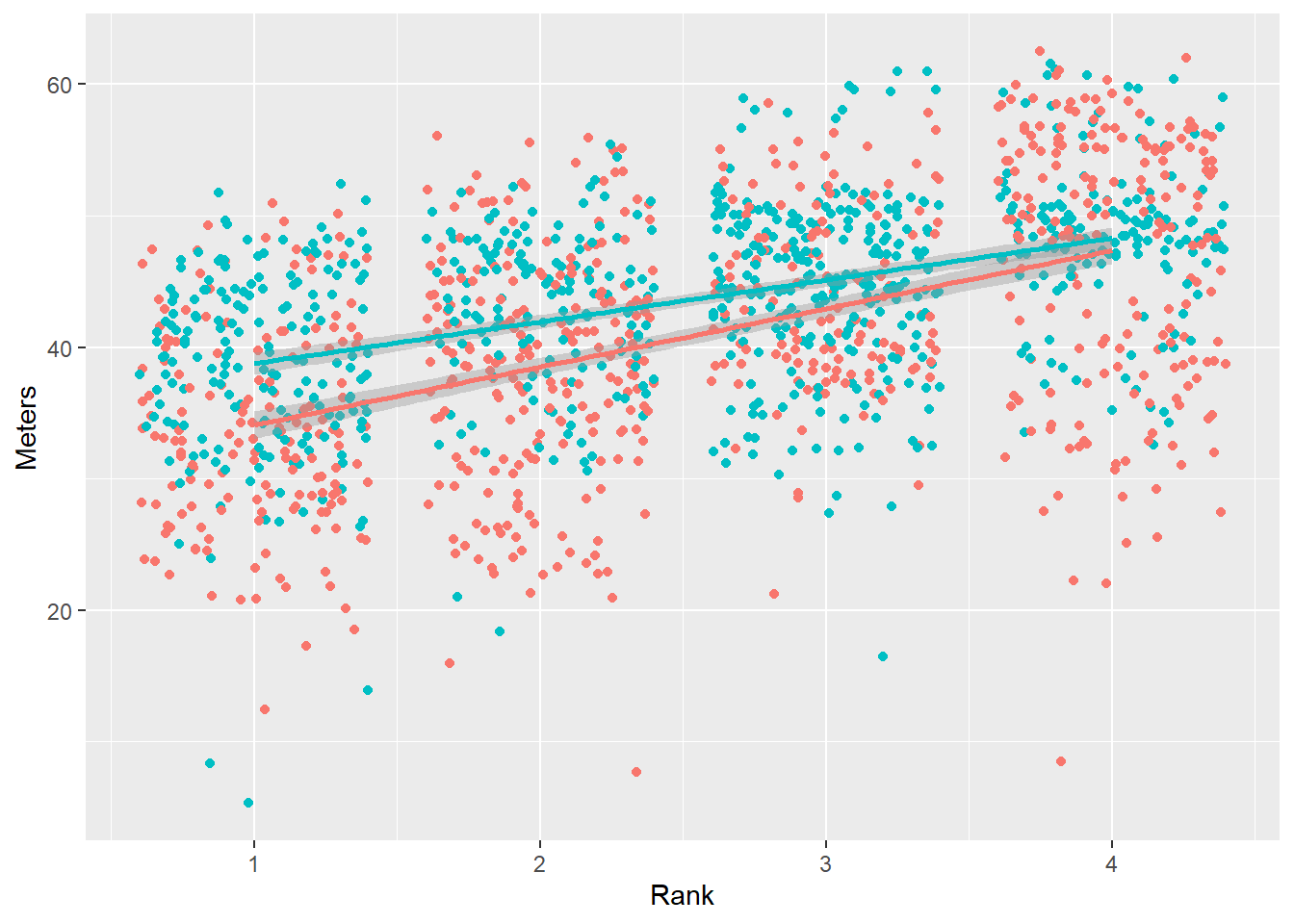
We see an overall pattern in the data. We can see a pattern for the Team subgroups across Rank.
We can add a facet_wrap() layer in order to plot the teams separately.
base_plot +
facet_wrap(facets = ~Team)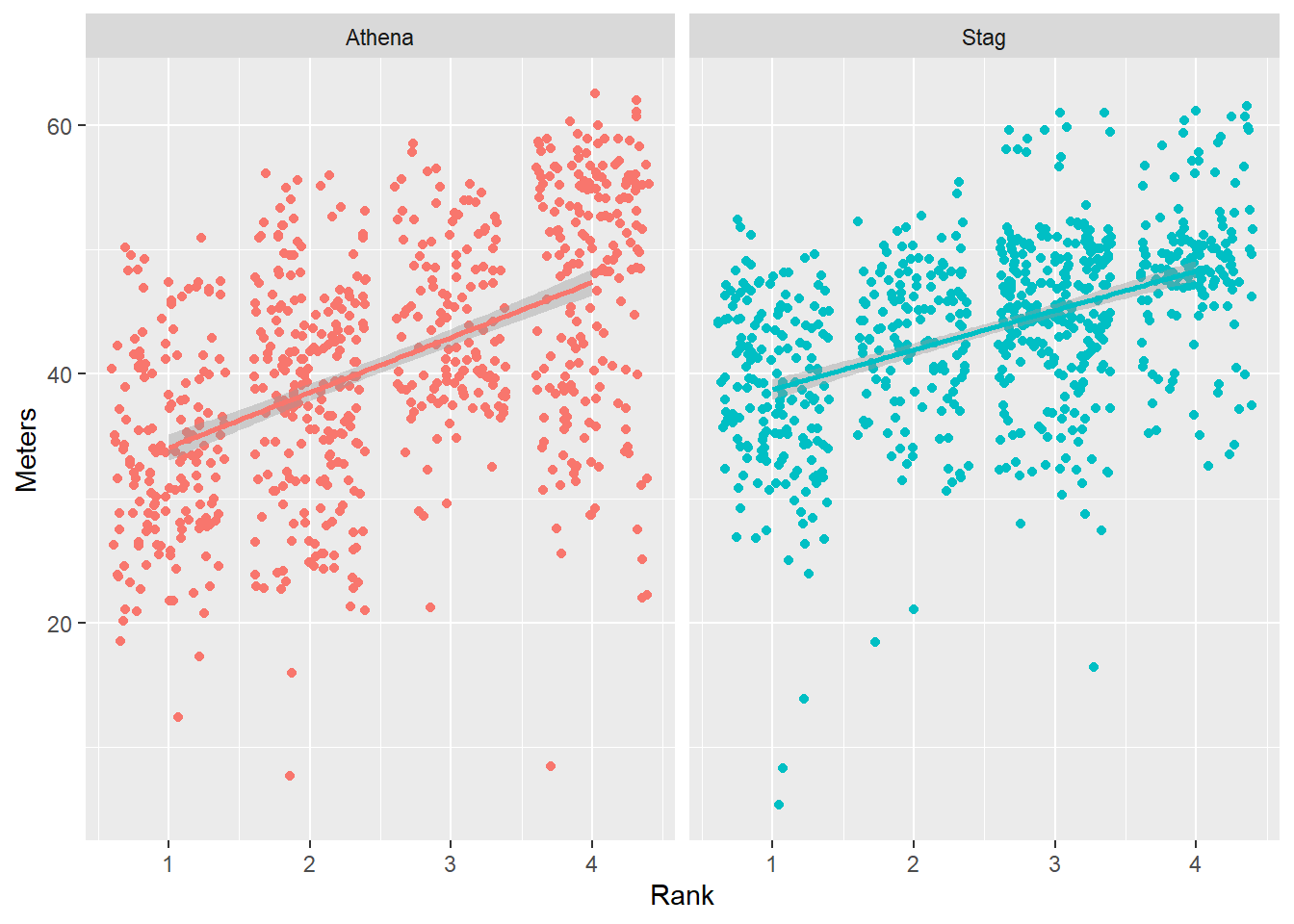
We can add a facet_wrap() layer in order to plot separately per year.
base_plot +
facet_wrap(facets = ~Year)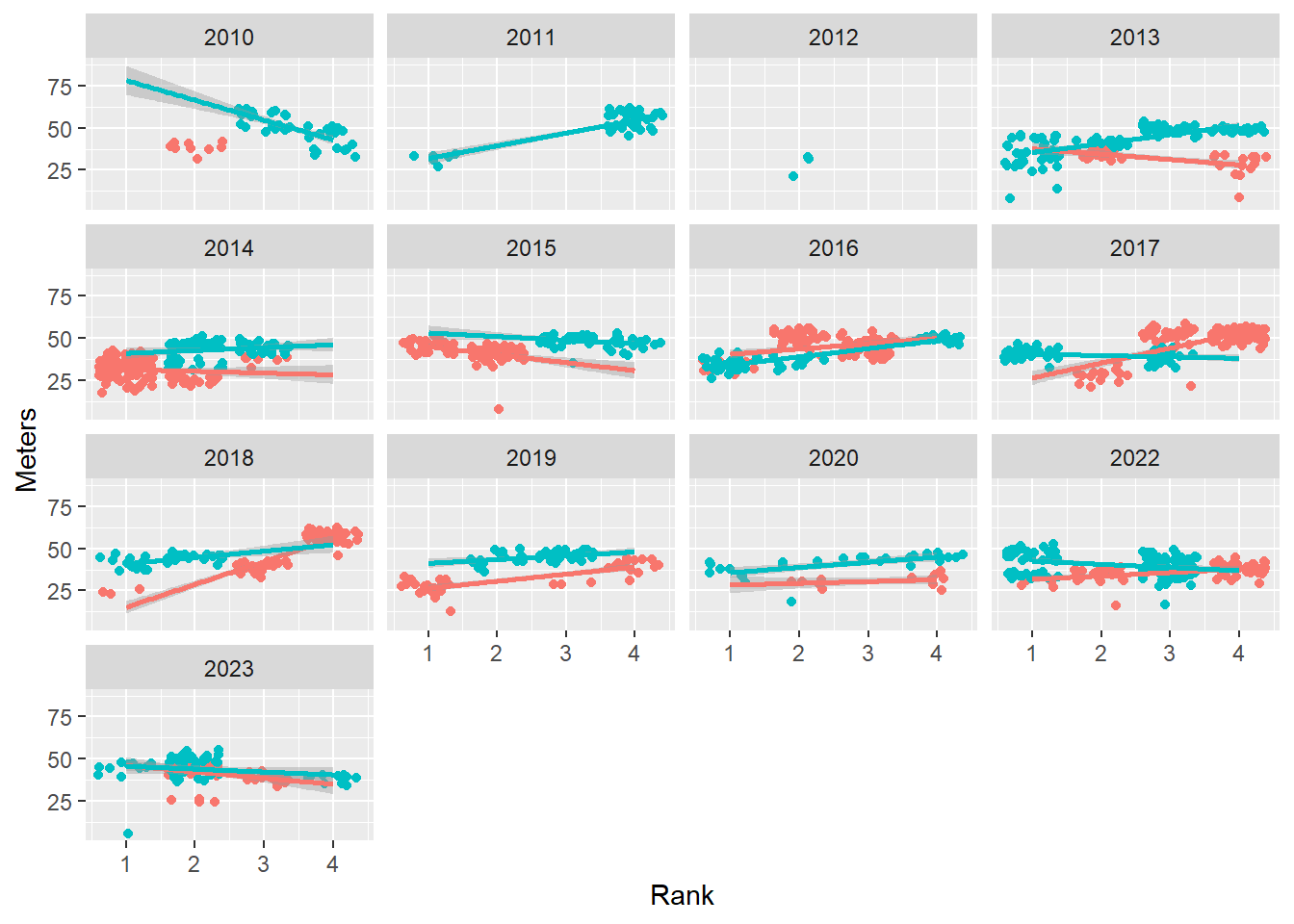
Notice that the x and y axis labels appear but the facet variable name appears as a label into the small multiples.
Creating facets using facet_wrap(facets = vars())
Use vars() instead of ~ to specify the variable because it will offer you greater flexibility and will reduce complications with faceting.
base_plot +
facet_wrap(facets = vars(Year))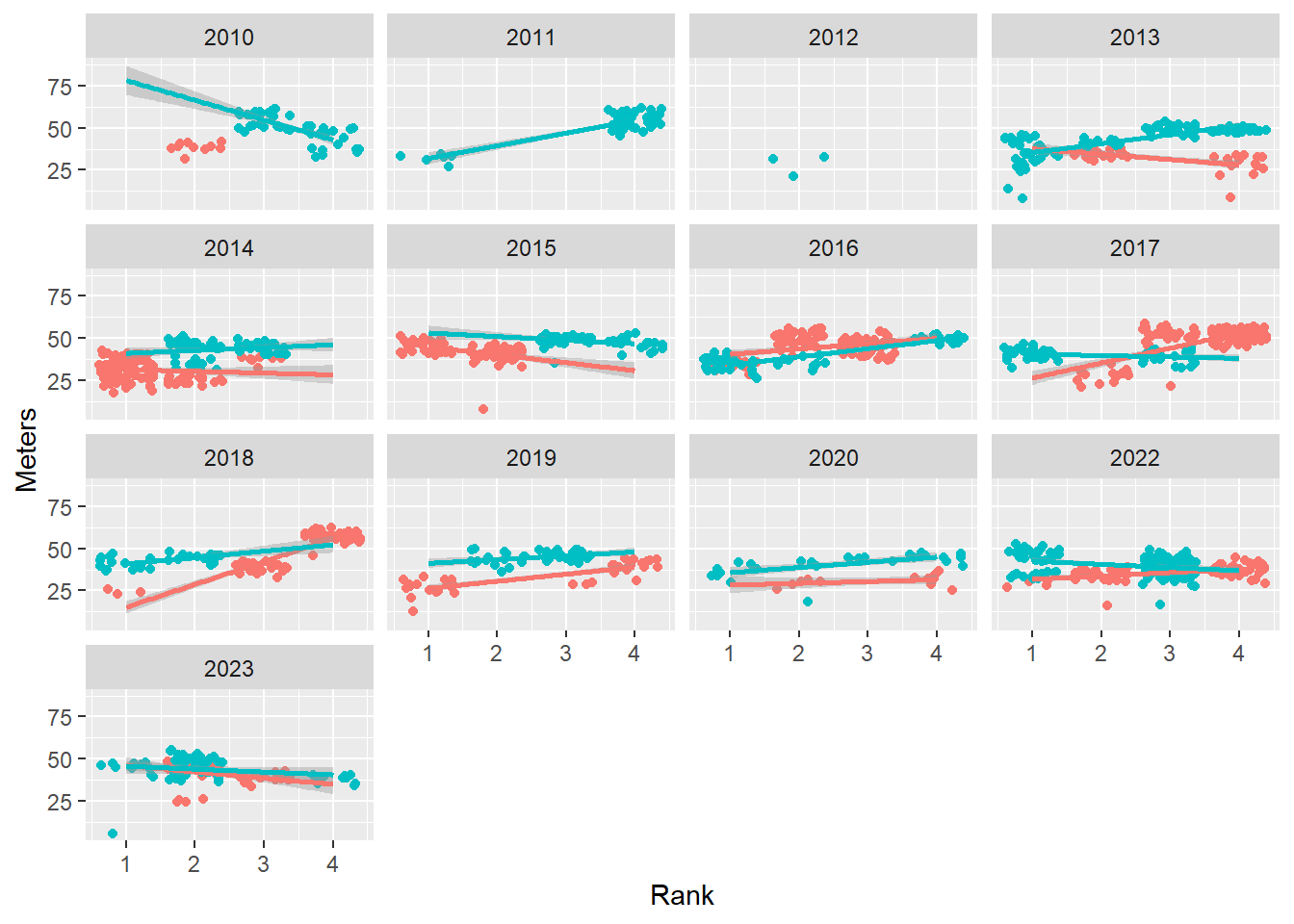
Passing two variables to vars() will create subplots for each.
base_plot +
facet_wrap(facets = vars(Year, Team))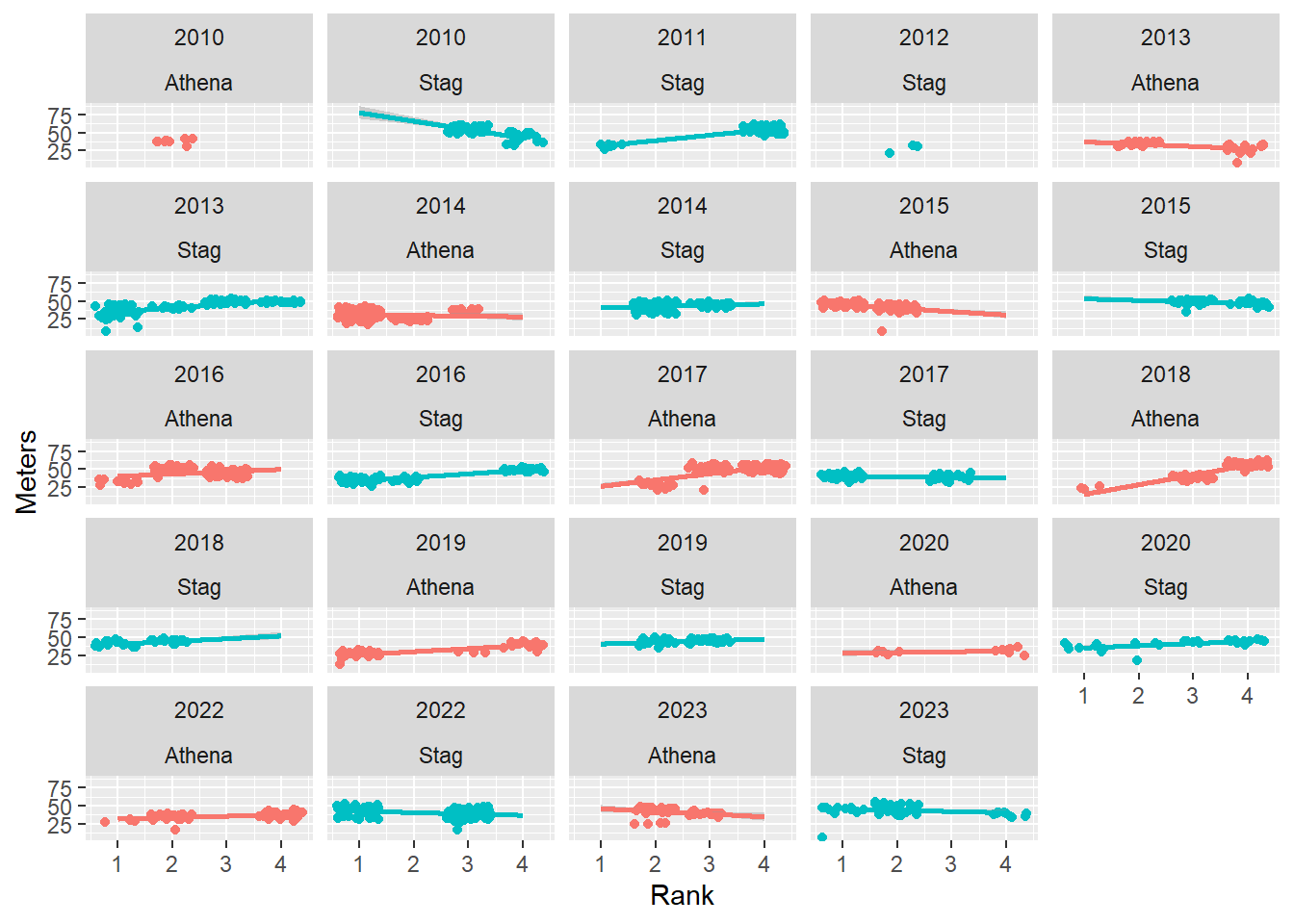
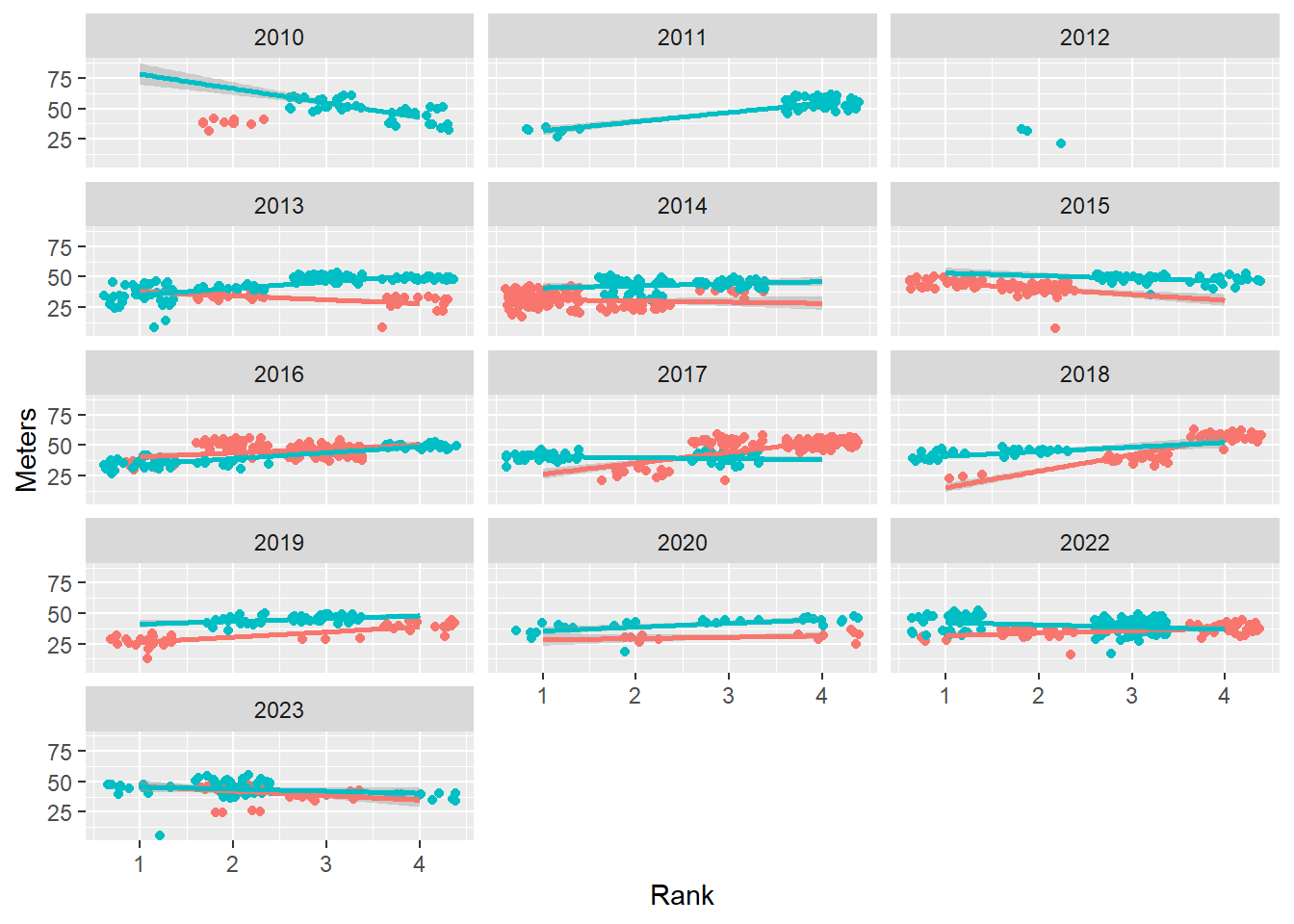
Controlling columns
Control the number of columns by setting ncol.
base_plot +
facet_wrap(facets = vars(Year),
ncol = 3
)
Controlling direction of arrangement
Change the direction using dir. By default, dir = "h" for a horizontal arrangement. The faceted variable changes from left to right, and top to bottom. Change to dir = "v".
base_plot +
facet_wrap(facets = vars(Year),
ncol = 3,
dir = "v"
)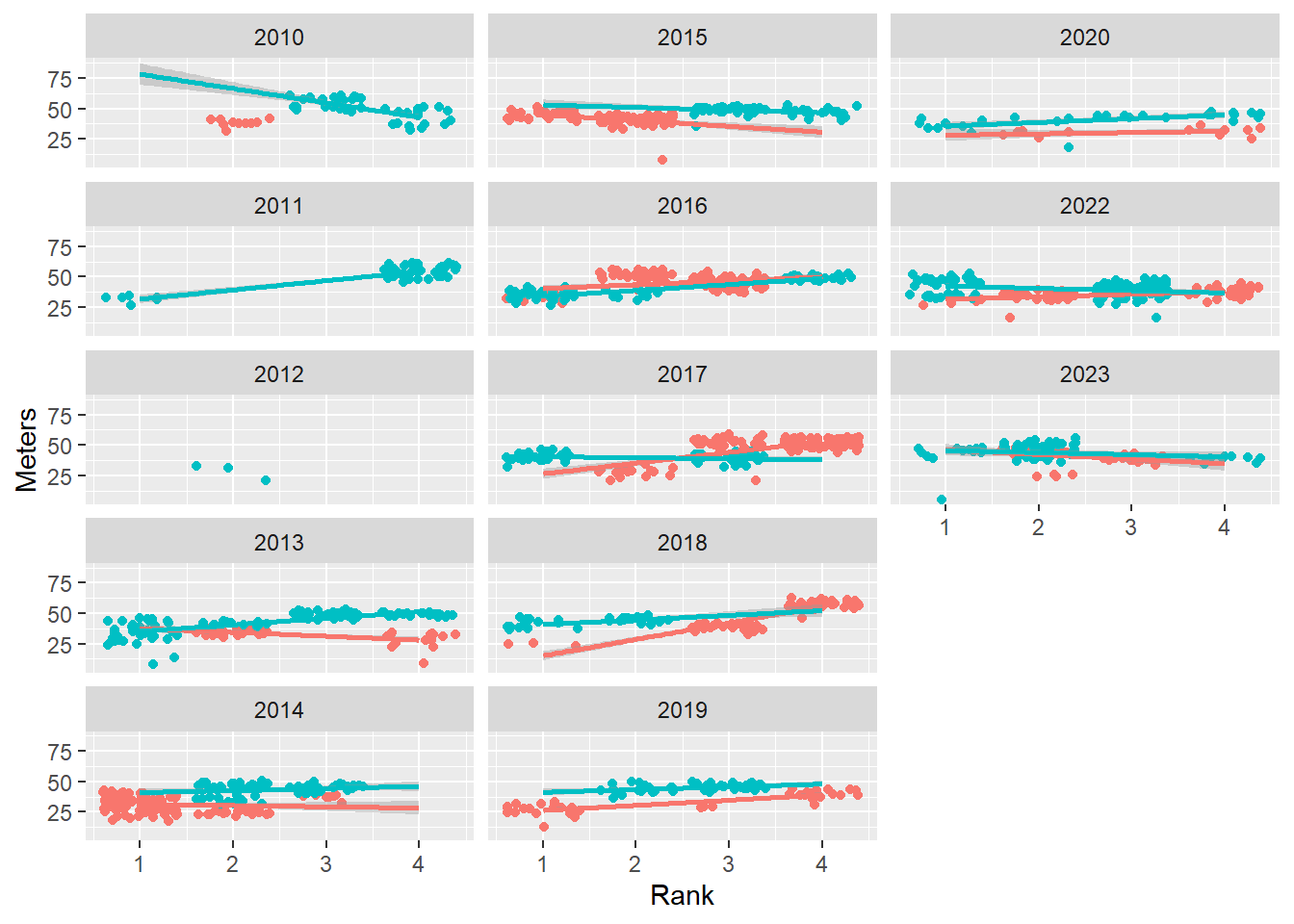
The faceted variable now changes from top to bottom, left to right. The arrangement may depend on your goals or how your audience will make comparisons.
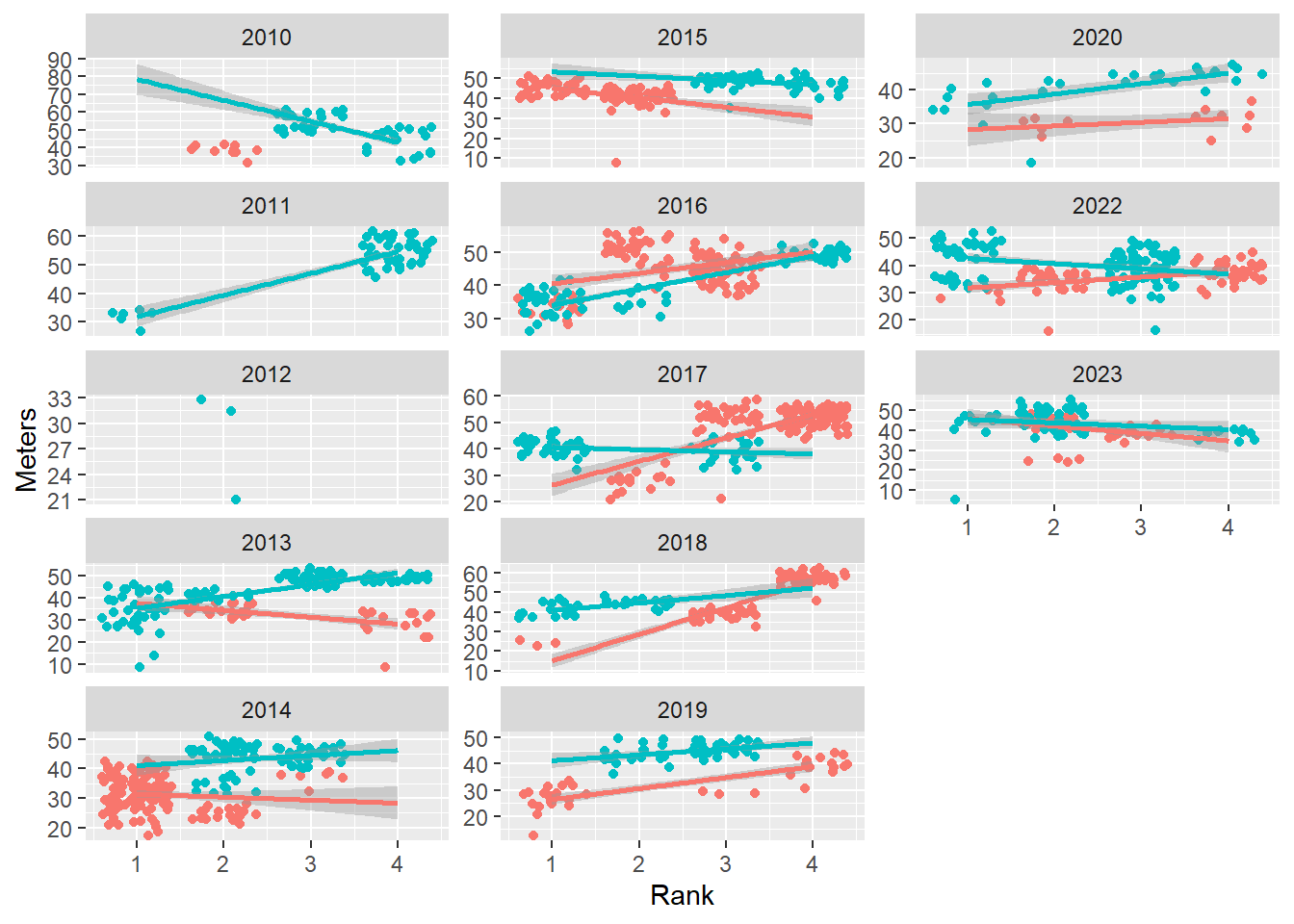
Controlling the scales
Speaking of comparisons being made, when the range of values for variables varies by facet level, you may wish to constrain the scales or allow them to vary. By default, scales = "fixed" but you can change to move freely for x, y, or both x and y.
scales = "fixed": fix both x and y scales to be the same across plots (default) scales = "free_x": x can vary freely, fix y scales = "free_y": y can vary freely, fix x scales = "free": x and y can vary freely
Allow scales = "free_y":
base_plot +
facet_wrap(facets = vars(Year),
ncol = 3,
dir = "v",
scales = "free_y"
)
Allow both scales = "free":
base_plot +
facet_wrap(facets = vars(Year),
ncol = 3,
dir = "v",
scales = "free"
)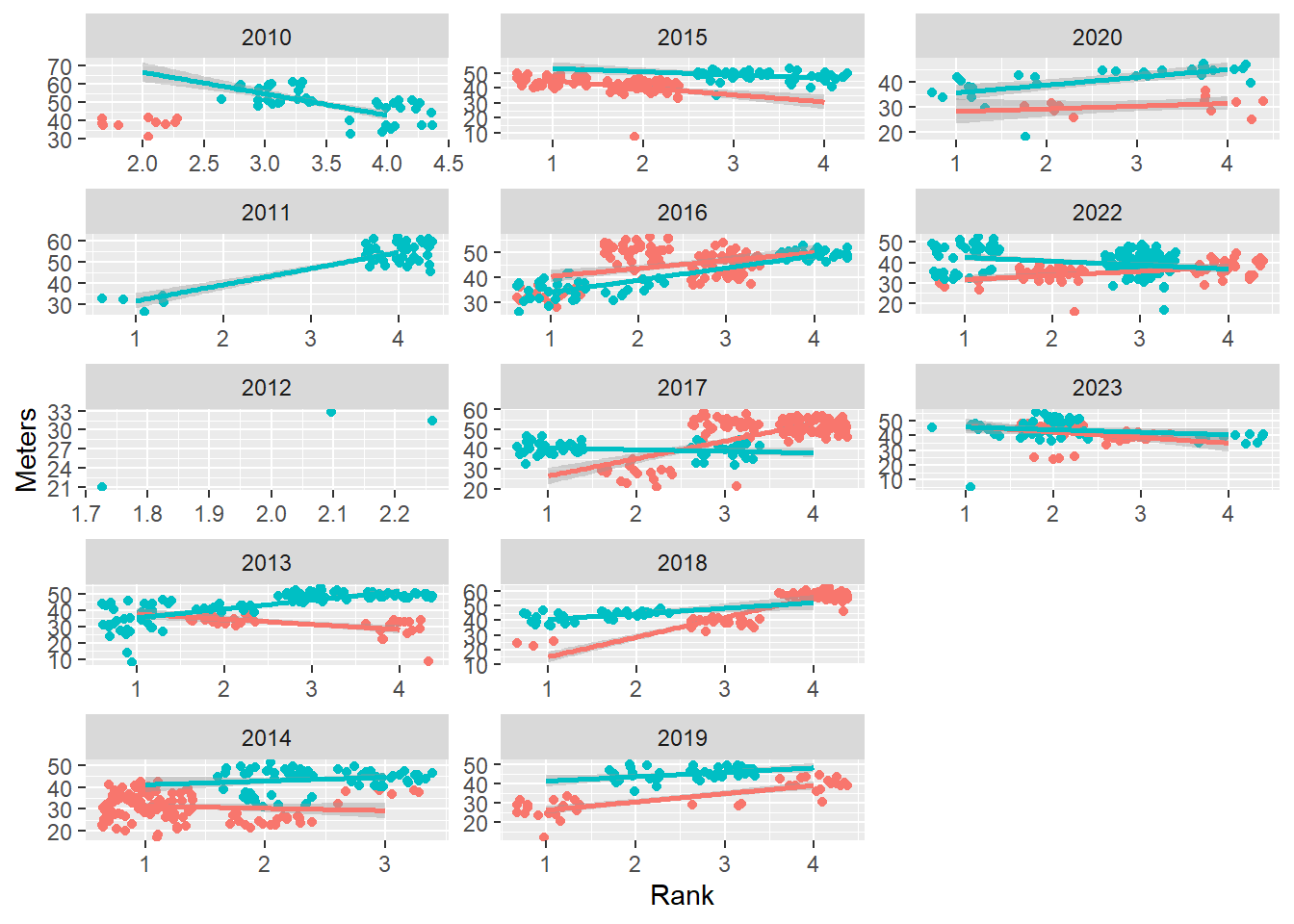
In this instance, comparing position of points is quite complicated because the axes are not aligned in order to support those comparisons. The view has to evaluate the axes, extract out the values, and then compare. Subtle patterns may be easier to see, however.
Ordering facets
The default ordering of facets is alphabetic/numeric. This may be fine when your facet variable is numeric but it may not be helpful when your facet variable is of character type. When your facet variable is not ordered as you wish, you can change the over by making your variable a factor() and specify the level order.
The default base plot is:
(base_plot <-
DATA |>
filter(!is.na(Rank)) |>
ggplot(mapping = aes(x = Rank,
y = Meters,
color = Team
)) +
geom_point(position = position_jitter()) +
geom_smooth(mapping = aes(color = Team),
method = "lm",
fullrange = TRUE
) +
theme(legend.position = "none")
)
DATA |>
ggplot(mapping = aes(x = Rank,
y = Meters,
)) +
geom_point(position = position_jitter()) +
geom_smooth(mapping = aes(color = Team),
method = "lm",
fullrange = TRUE
) +
theme(legend.position = "none") +
facet_wrap(facets = vars(Team))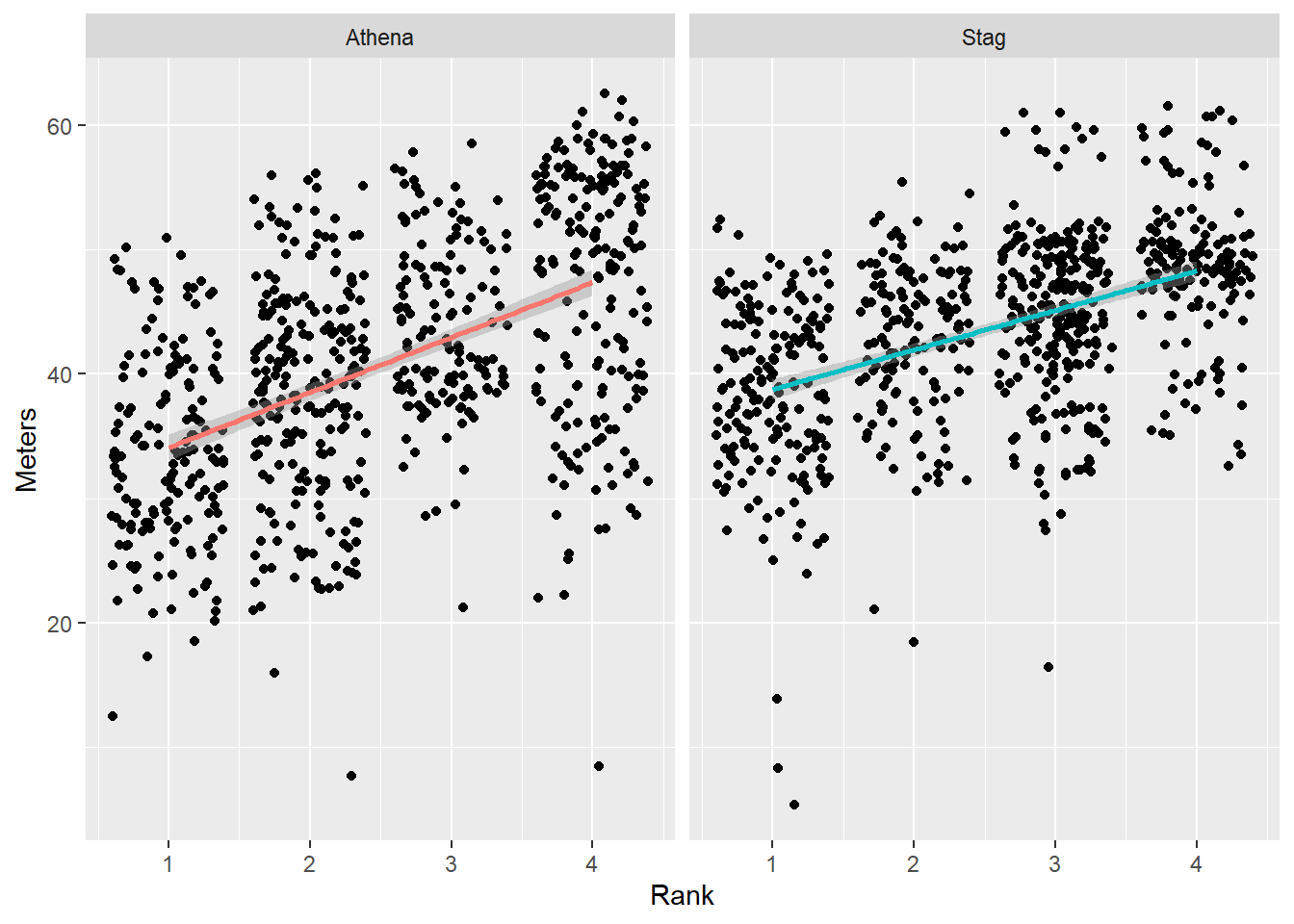
Change the order by ordering the factor()
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team,
levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
#color = Team
)) +
geom_point(position = position_jitter()) +
geom_smooth(method = "lm") +
theme(legend.position = "none") +
facet_wrap(facets = vars(Team))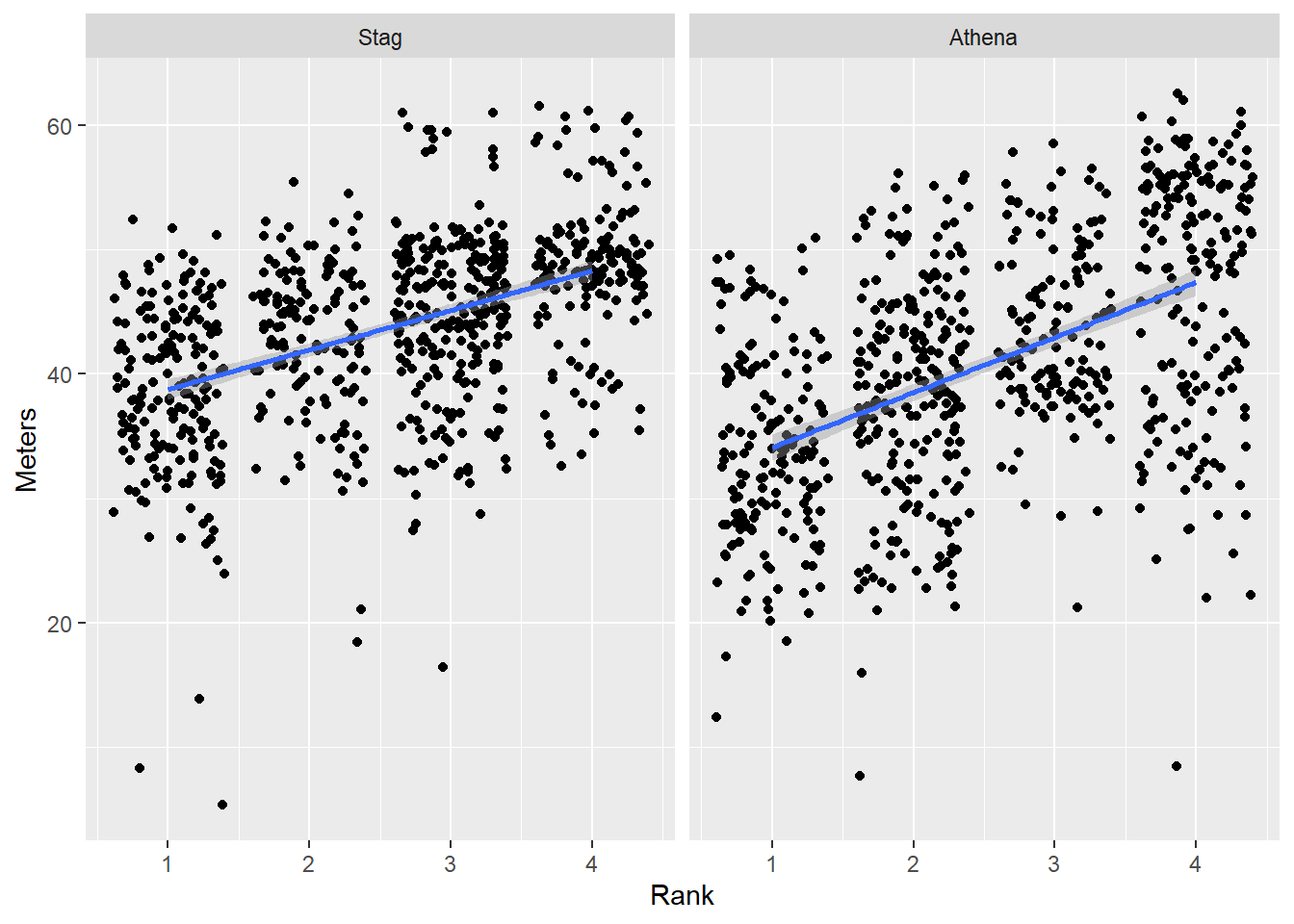
Change the order by ordering of the data using forcats::fct_reorder()
We can use forcats::fct_reorder() for reordering as we have done before for single plots. When the levels of a factor occur more than once, fct_reorder() applies a summary function, which by default is median() and the sorting order is from lowest to highest value.
Order by the default behavior
We can add the arguments so they are visible.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team,
levels = c("Stag", "Athena"))) |>
mutate(Team = forcats::fct_reorder(.f = Team, # the factor to sort
.x = Meters, # the variable to sort by
.fun = median, # the default function
.desc = FALSE # the default sorting behavior
)
) |>
ggplot(mapping = aes(x = Rank,
y = Meters,
)) +
geom_point(position = position_jitter()) +
geom_smooth(method = "lm") +
theme(legend.position = "none") +
facet_wrap(facets = vars(Team))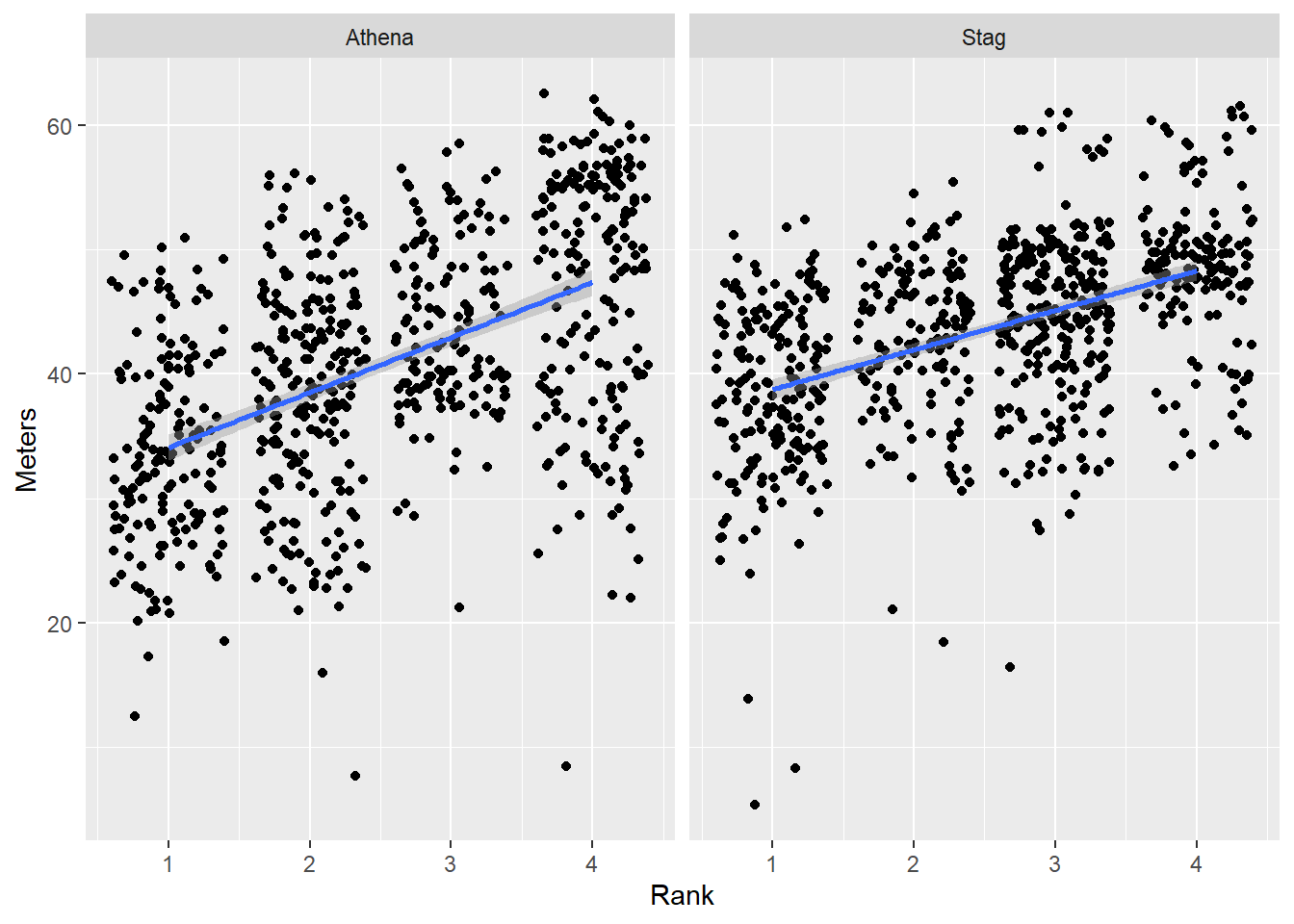
Order by the mean() and sort from highest to lowest
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = forcats::fct_reorder(.f = Team, # the factor to sort
.x = Meters, # the variable to sort by
.fun = mean, # the mean function
.desc = FALSE # the default sorting behavior
)
) |>
ggplot(mapping = aes(x = Rank,
y = Meters,
)) +
geom_point(position = position_jitter()) +
geom_smooth(method = "lm") +
theme(legend.position = "none") +
facet_wrap(facets = vars(Team))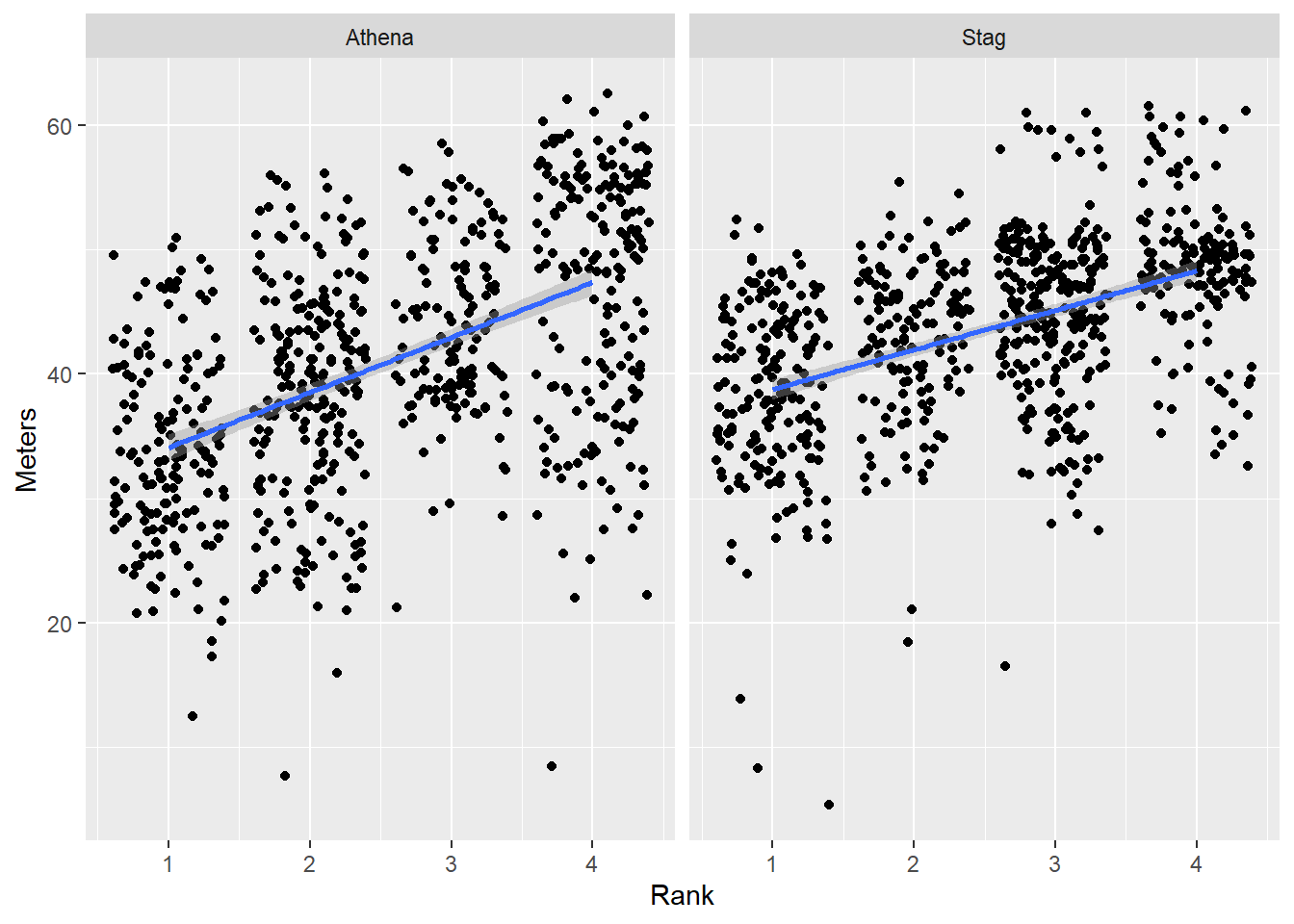
Order by a function
We can pass a function that determines the difference between the mean() and the median(). If we can about the difference being positive or negative, the order of these computations will matter. If we want to order based on the size of the difference between the two metrics, we can take the absolute value of the difference using abs().
~ abs(med(.x) - mean(.x)): a difference between median and meanmean(x, trim = 0.1): the mean that is based on trimming the outlying 10%max(x) - min(x)): the range
DATA |>
mutate(Team = forcats::fct_reorder(.f = Team, # the factor to sort
.x = Meters, # the variable to sort by
# a function
.fun = ~ abs(median(.x) - mean(.x)),
# also but more complicated function approach
#.fun = function(x) { abs(median(x) - mean(x)) },
.desc = TRUE # from largest value to the smallest
)
) |>
ggplot(mapping = aes(x = Rank,
y = Meters,
)) +
geom_point(position = position_jitter()) +
geom_smooth(method = "lm") +
theme(legend.position = "none") +
facet_wrap(facets = vars(Team)) +
labs(title = NULL)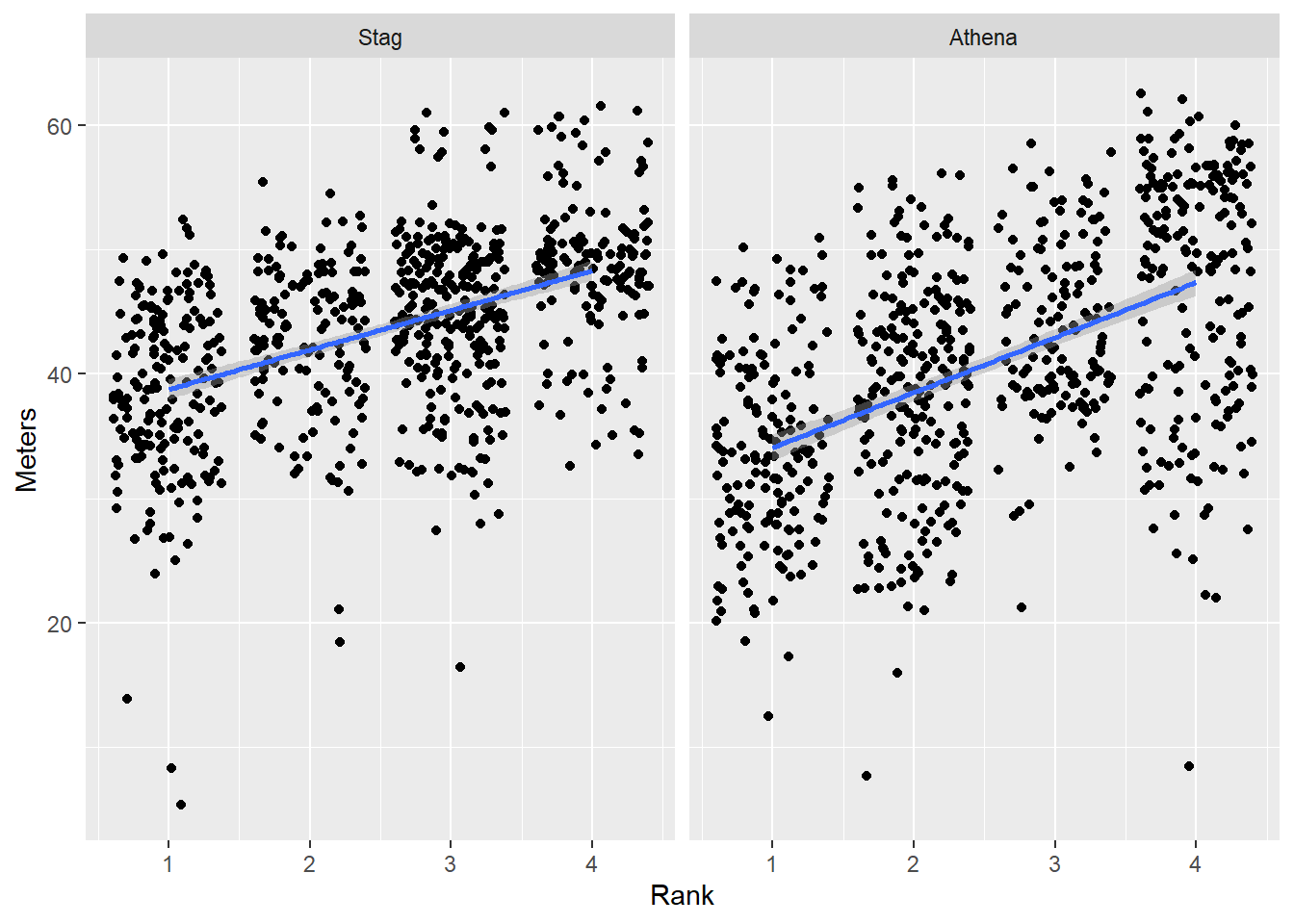
Importantly, the ordering of the panels in the facet plot always communicates information that is not visible or easily extracted from the visualization. By default, this ordering is by character or number. The facet with the largest mean may appear in facet position 1 or 12 (think random) but your goal may not be to communicate that difference across facets. A systematic ordering of the panels, however, using fct_reorder() represents a decision (whether conscious or not) to order the panels based on some method other than the default alphabetical or numeric order. This consequence is because both fct_reorder() and fct_reorder2() apply a function by with to order the data based on some variable and they then apply a sorting method based on that function. When you consciously apply fct_reorder(), you are doing so for a specific reason. Thus, you would want to communicate that information either in the plot title, subtitle, caption, or in a written report. Keep in mind that the ordering of panels reflects data compared across panels in the data visualization.
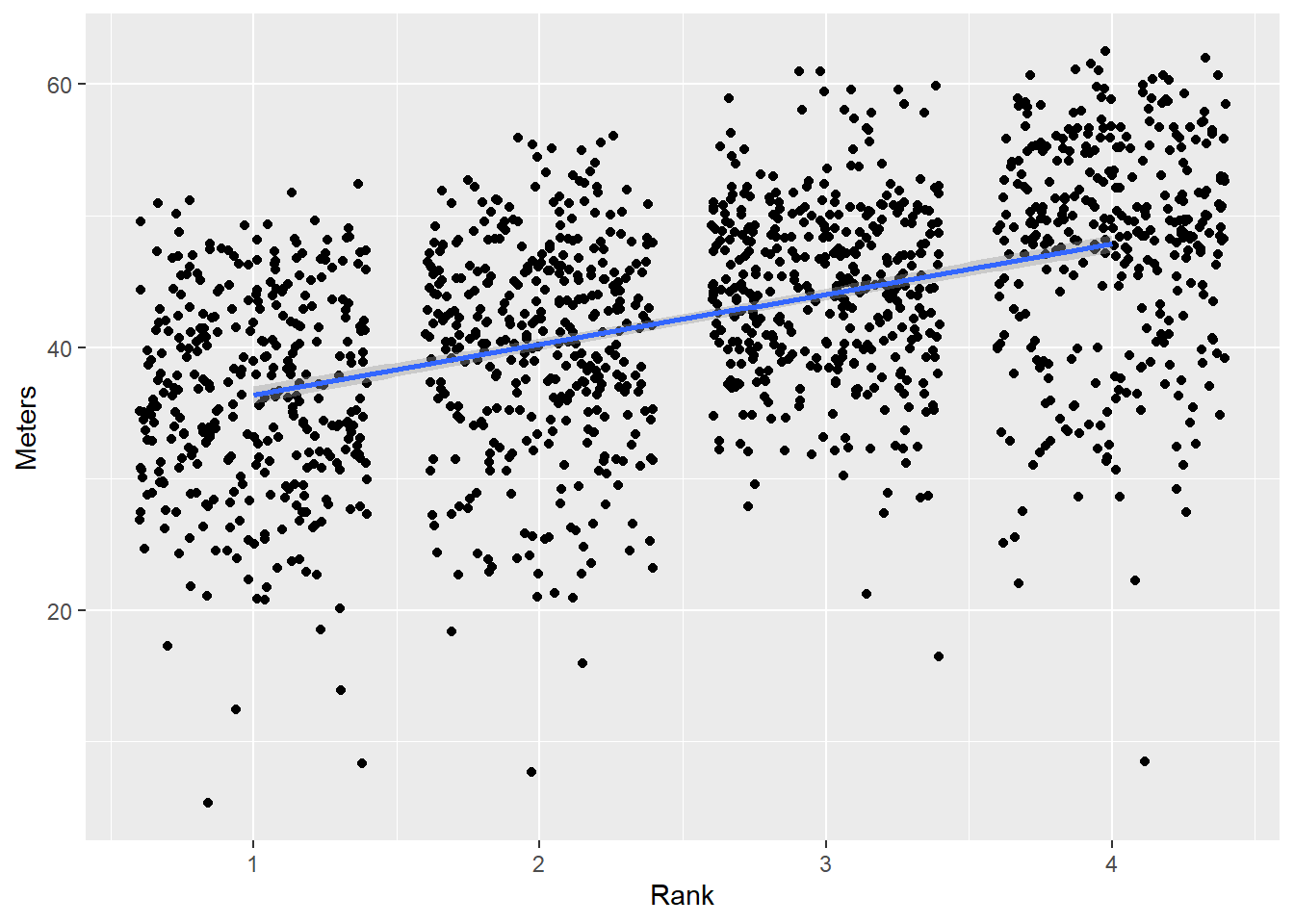
Repositioning the label strip
By default, the facet label will be on the top of the plot because strip.position = "top" but you can set to "top", "bottom", "left", or "right".
Let’s set strip.position = "left":
(base_plot <-
DATA |>
mutate(Team = forcats::fct_reorder(.f = Team, # the factor to sort
.x = Meters, # the variable to sort by
.fun = mean, # the mean function
.desc = FALSE # the default sorting behavior
)
) |>
ggplot(mapping = aes(x = Rank,
y = Meters,
)) +
geom_point(position = position_jitter()) +
geom_smooth(method = "lm") +
theme(legend.position = "none")
)
base_plot +
facet_wrap(facets = vars(Team),
strip.position = "left"
)
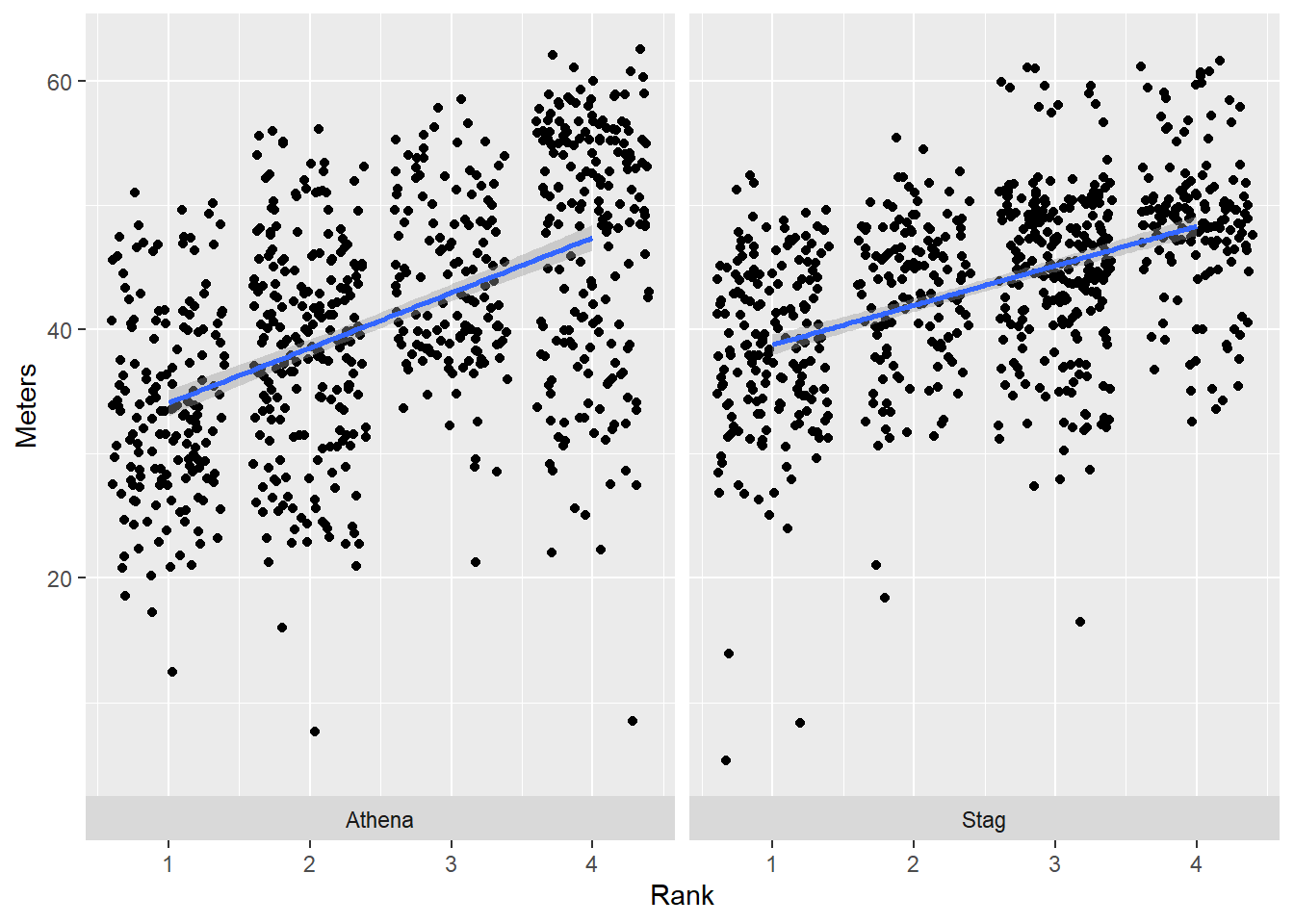
Let’s set strip.position = "bottom":
base_plot +
facet_wrap(facets = vars(Team),
strip.position = "bottom"
)
Although this repositioning is handled by theme(), theme(strip.placement = "outside")
base_plot +
facet_wrap(facets = vars(Team),
strip.position = "bottom"
) +
theme(strip.placement = "outside")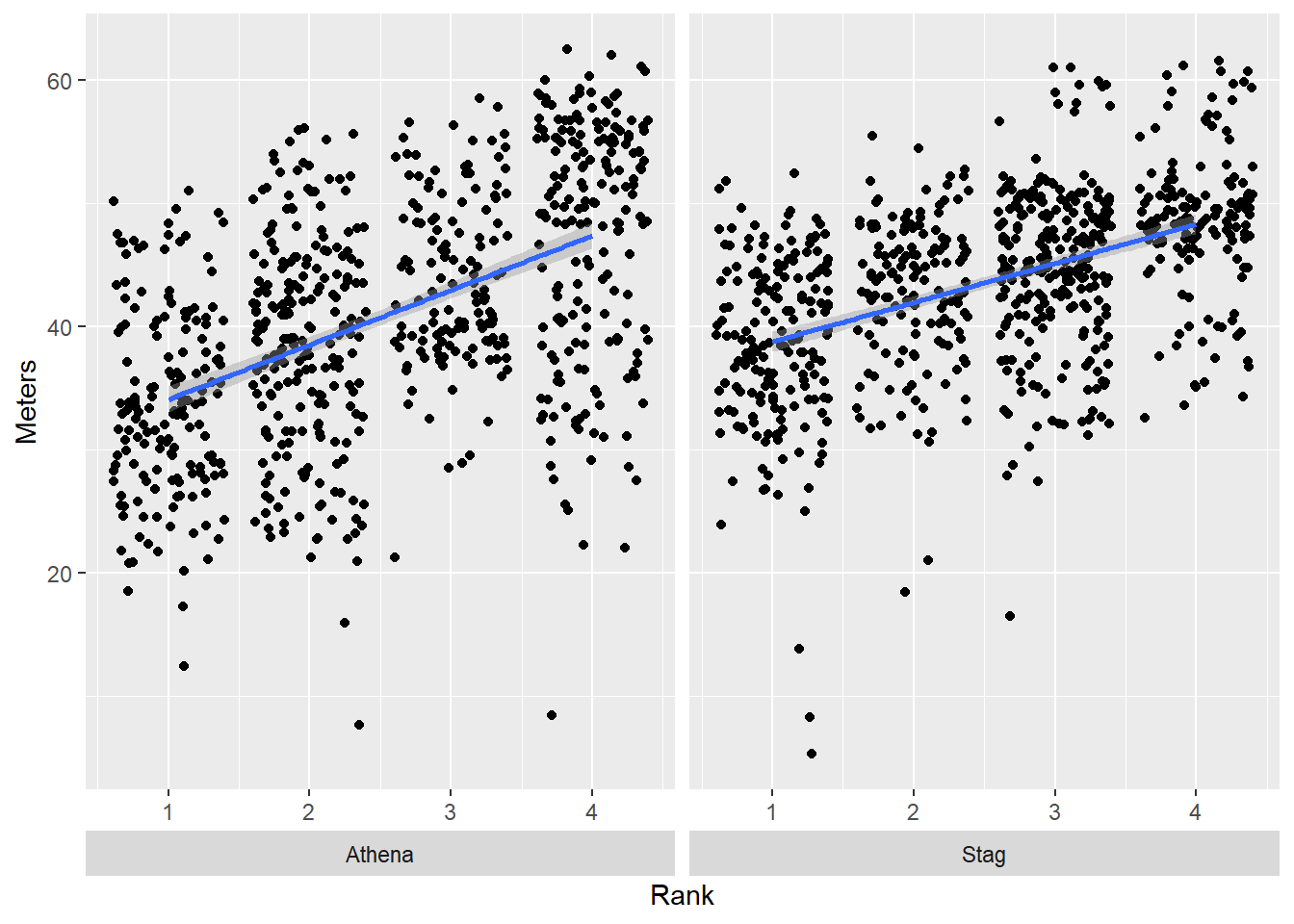
Faceting bars
Bar plots are faceted in the same manner as for points. The comparisons are just different, for example, with the height of bars.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = Team
)) +
stat_summary(fun = mean, geom = "bar")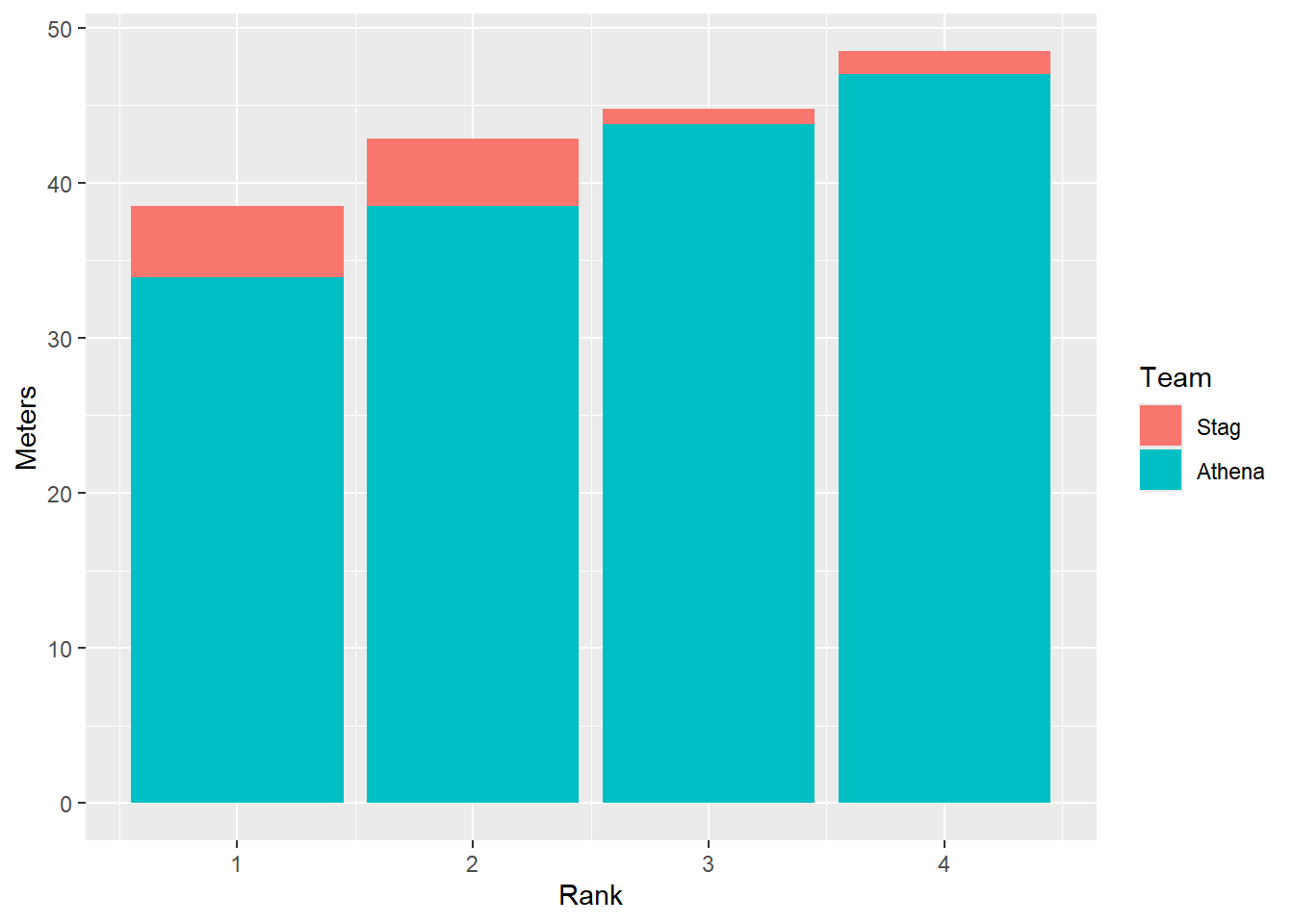
Remember, by default geom_col() will stack bars.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = Team
)) +
stat_summary(fun = mean, geom = "bar") +
facet_wrap(facet = vars(Team))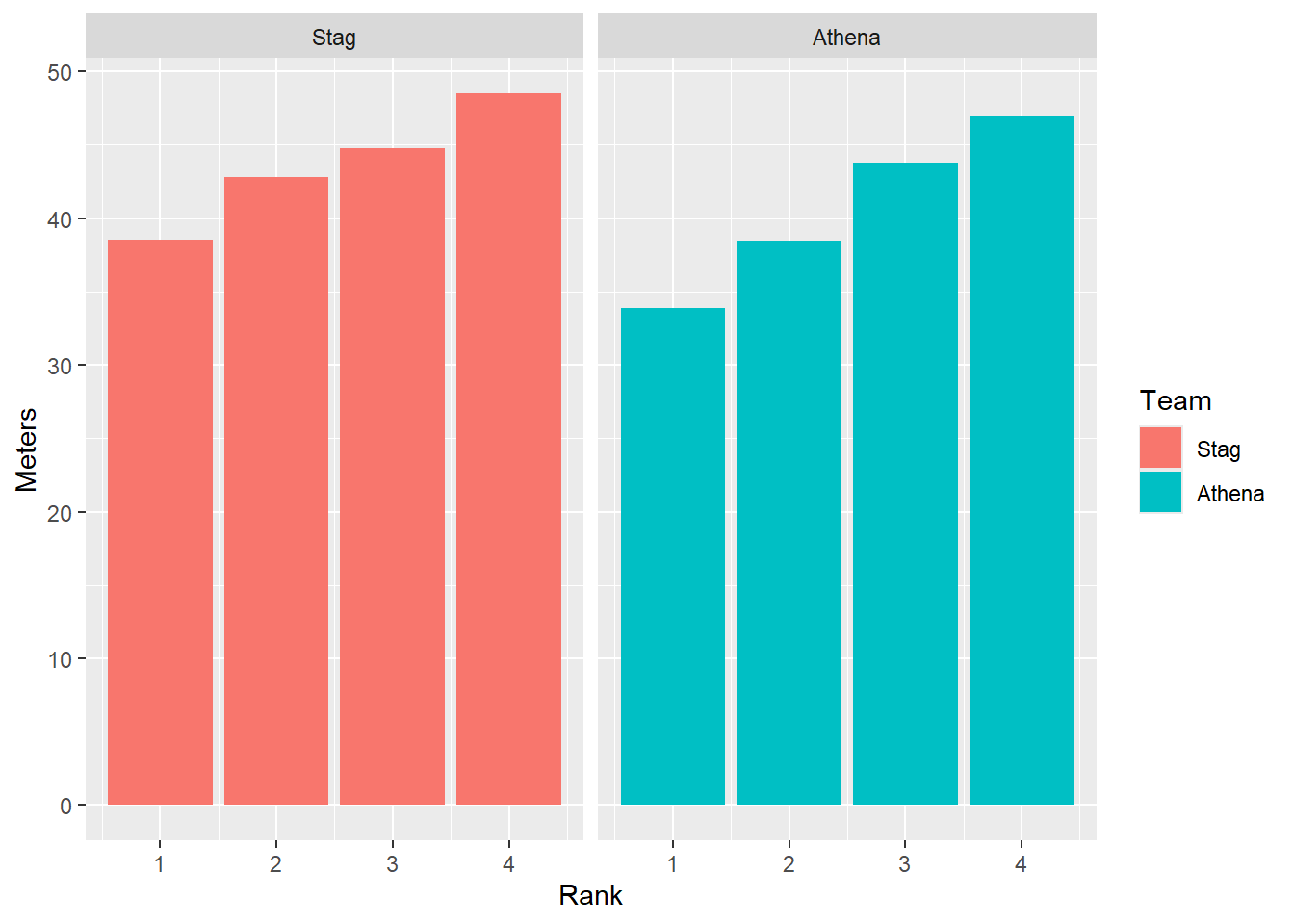
To remove any associate color that could be distracting, remove the aesthetic.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters
)) +
stat_summary(fun = mean, geom = "bar") +
facet_wrap(facet = vars(Team))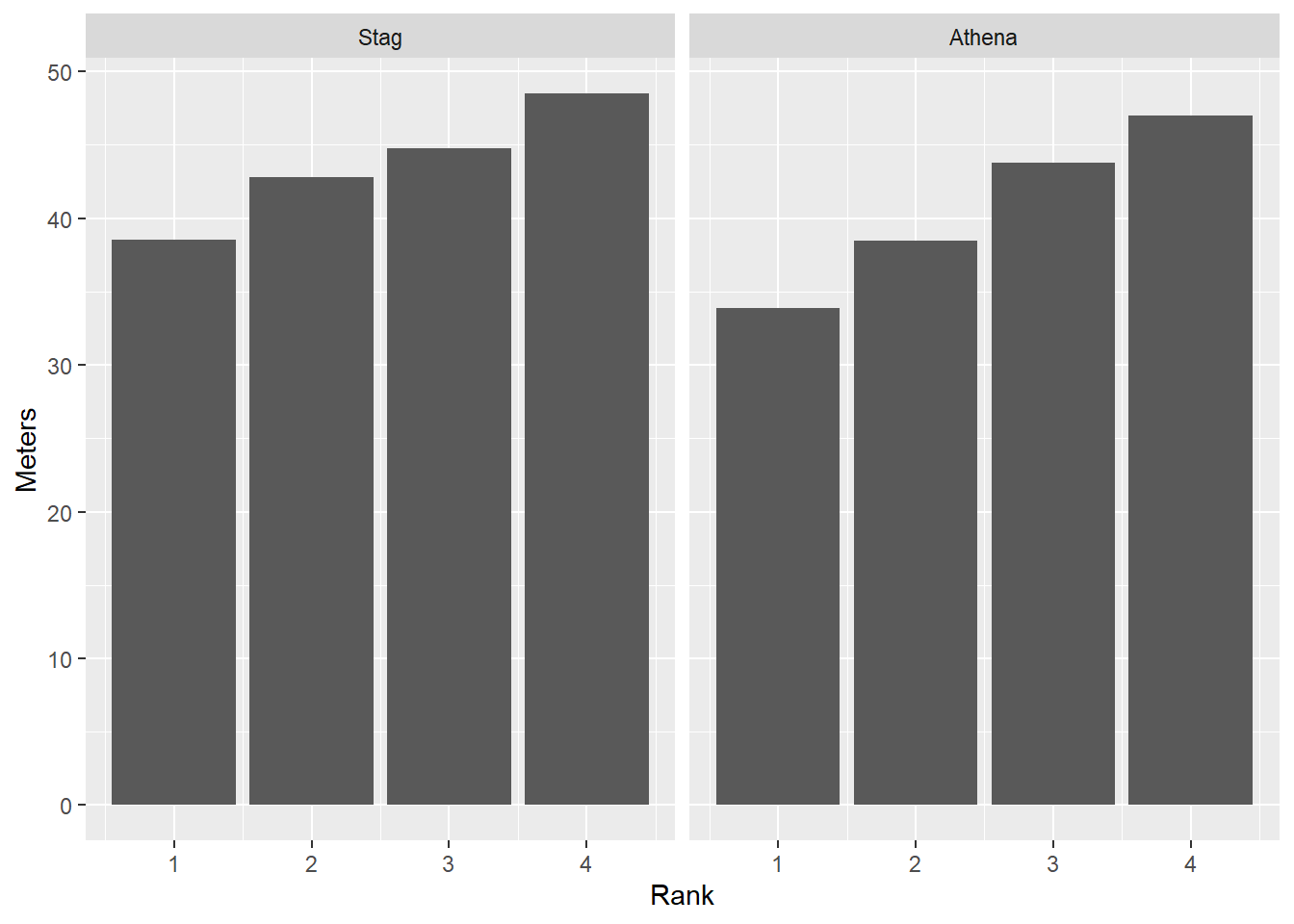
Or to facilitate comparisons across Rank, map that variable to fill.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = as.character(Rank) # or just Rank for continuous color bar
)) +
stat_summary(fun = mean, geom = "bar") +
facet_wrap(facet = vars(Team))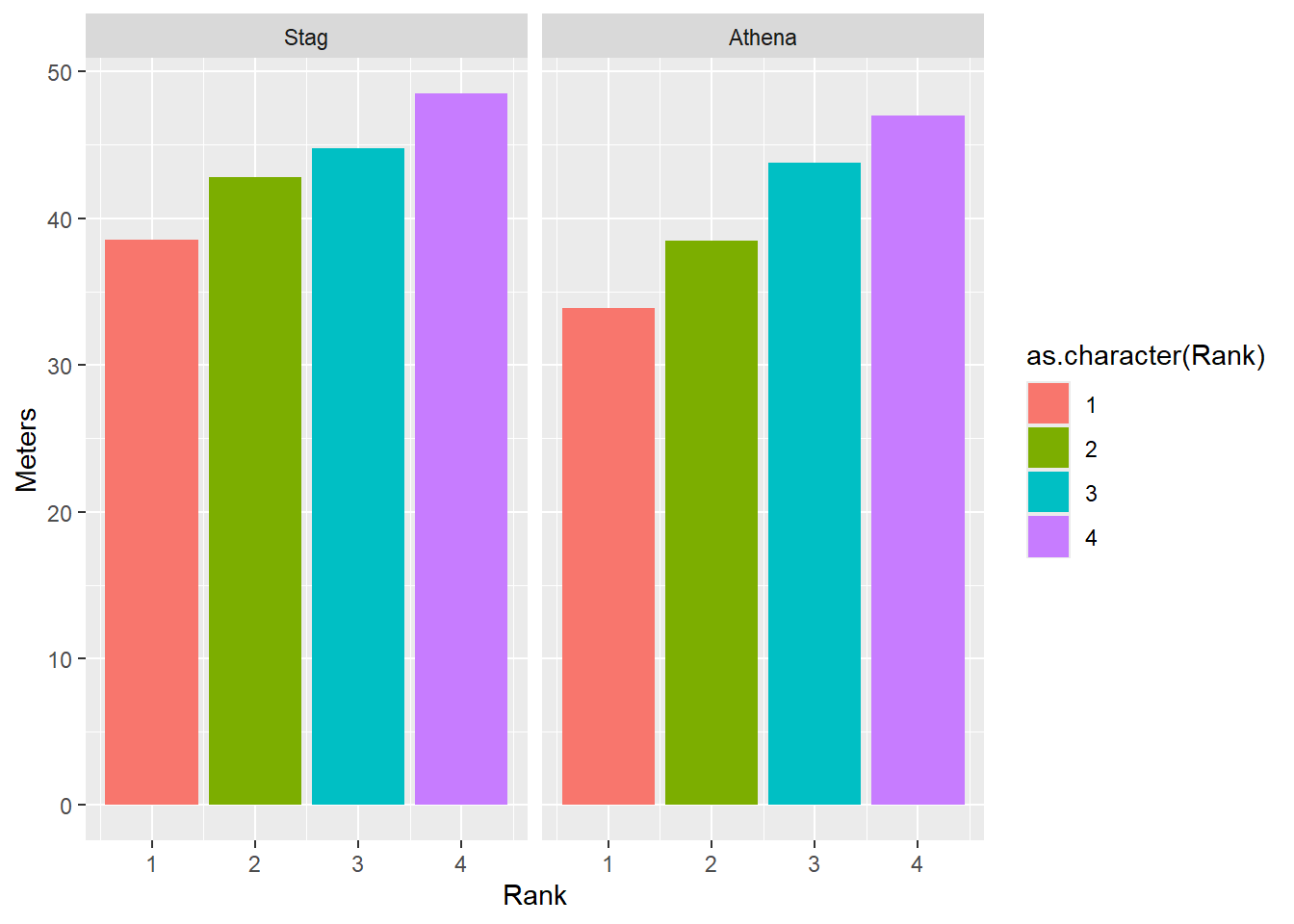
Arrangement matters. Comparing bars across unaligned rows (below) is more demanding cognitively.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = as.character(Rank) # or just Rank for continuous color bar
)) +
stat_summary(fun = mean, geom = "bar") +
facet_wrap(facet = vars(Team), ncol = 1)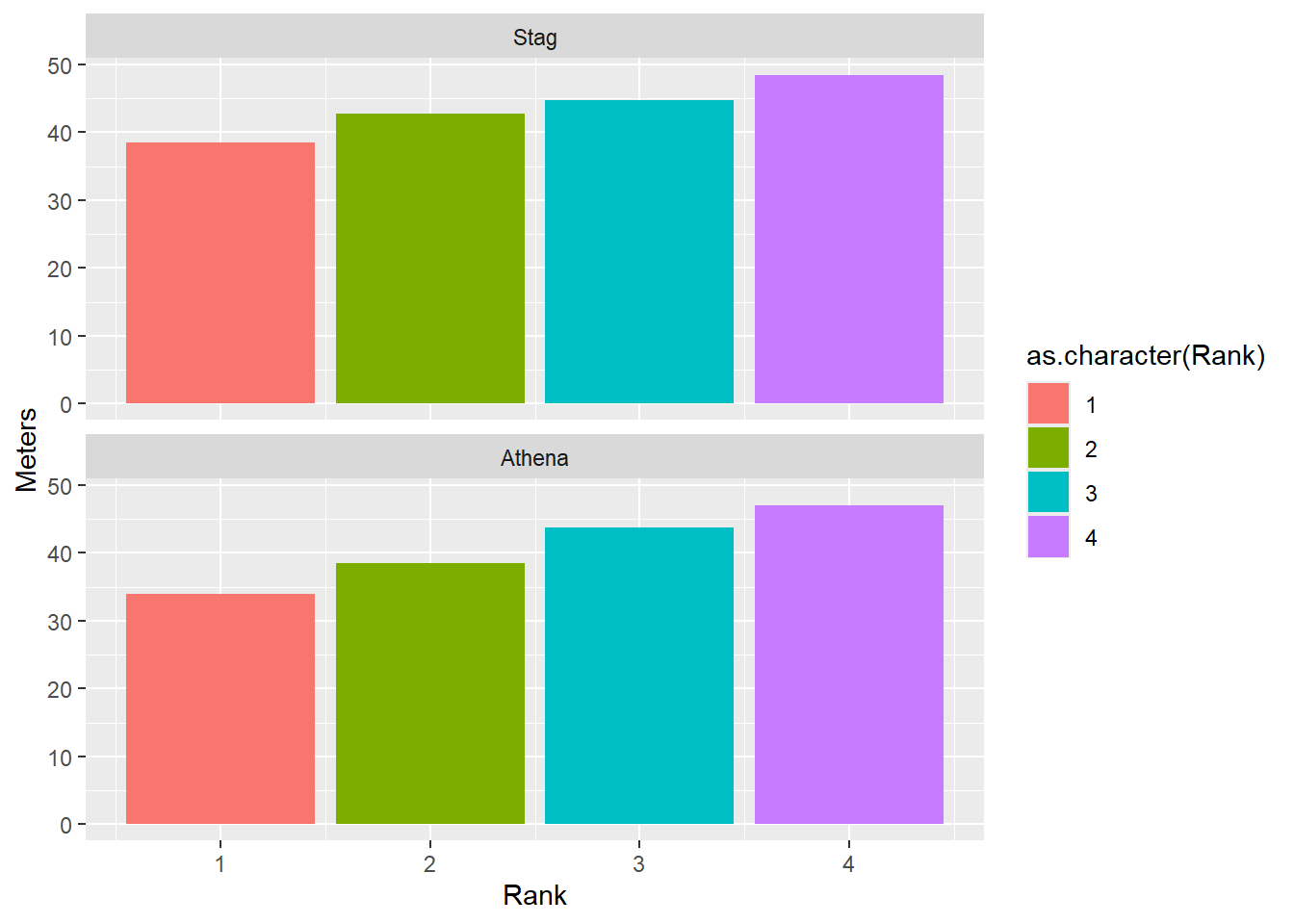
Facet Grid
Some of the same goals can be completed using facet_grid(). For this function, you will facet by specifying the rows and the cols for the grid.
facet_grid(rows = vars())
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = as.character(Rank) # or just Rank for continuous color bar
)) +
stat_summary(fun = mean, geom = "bar") +
facet_grid(rows = vars(Team))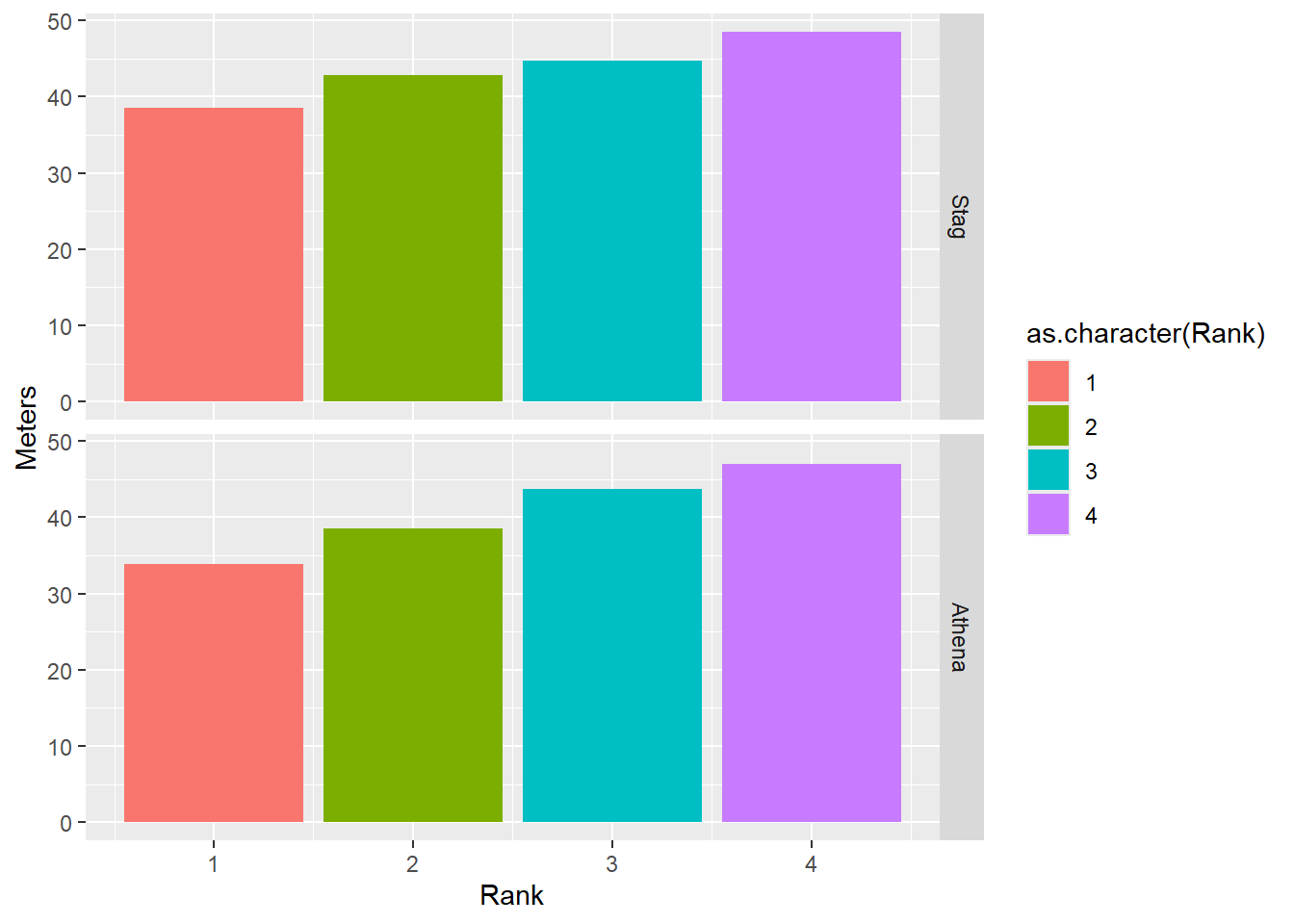
Be careful what you facet as you might create something you don’t intend.
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = as.character(Rank) # or just Rank for continuous color bar
)) +
stat_summary(fun = mean, geom = "bar") +
facet_grid(rows = vars(Rank))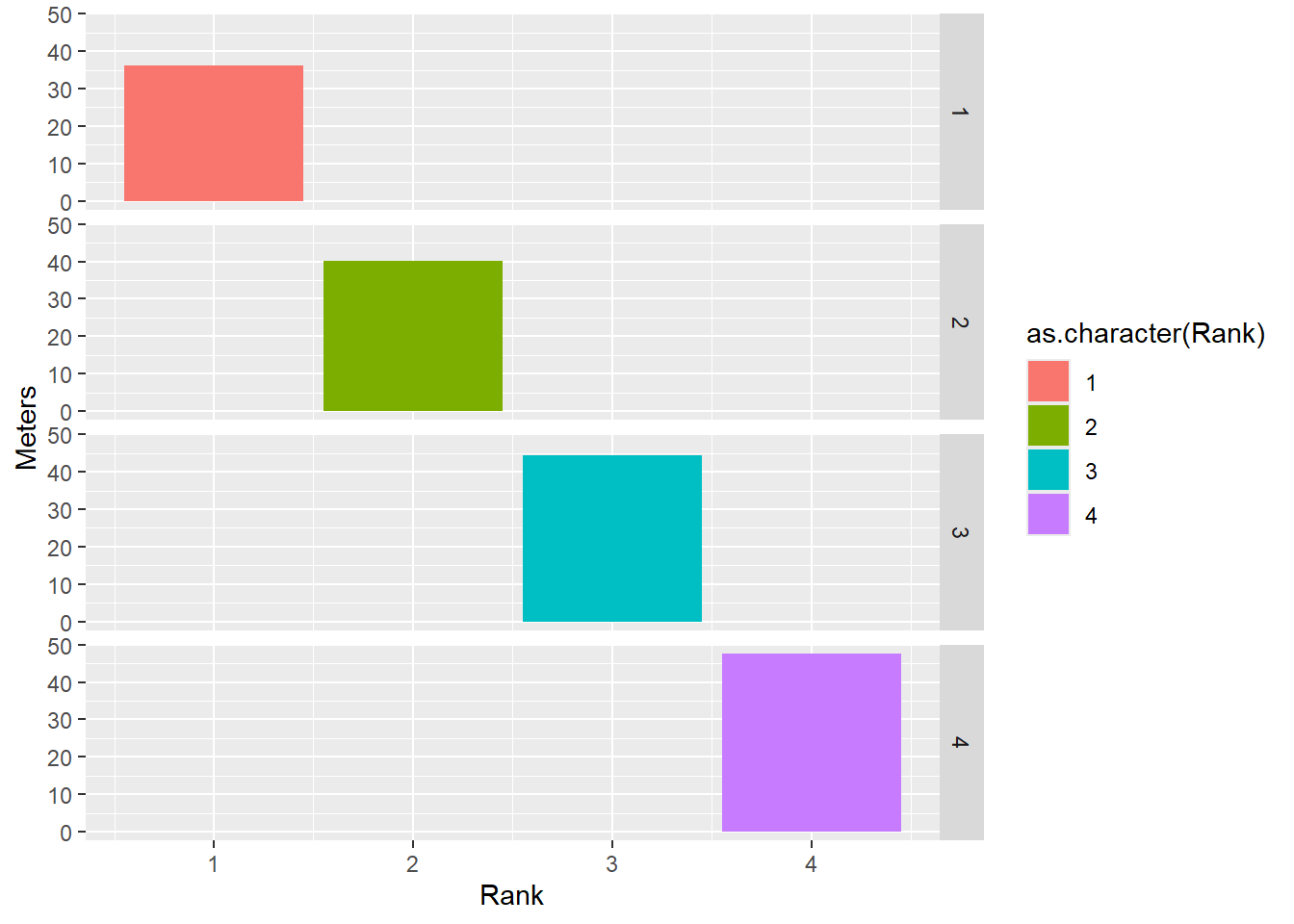
facet_grid(cols = vars())
DATA |>
filter(!is.na(Rank)) |>
mutate(Team = factor(Team, levels = c("Stag", "Athena"))) |> # factor and level order
ggplot(mapping = aes(x = Rank,
y = Meters,
fill = as.character(Rank) # or just Rank for continuous color bar
)) +
stat_summary(fun = mean, geom = "bar") +
facet_grid(cols = vars(Team))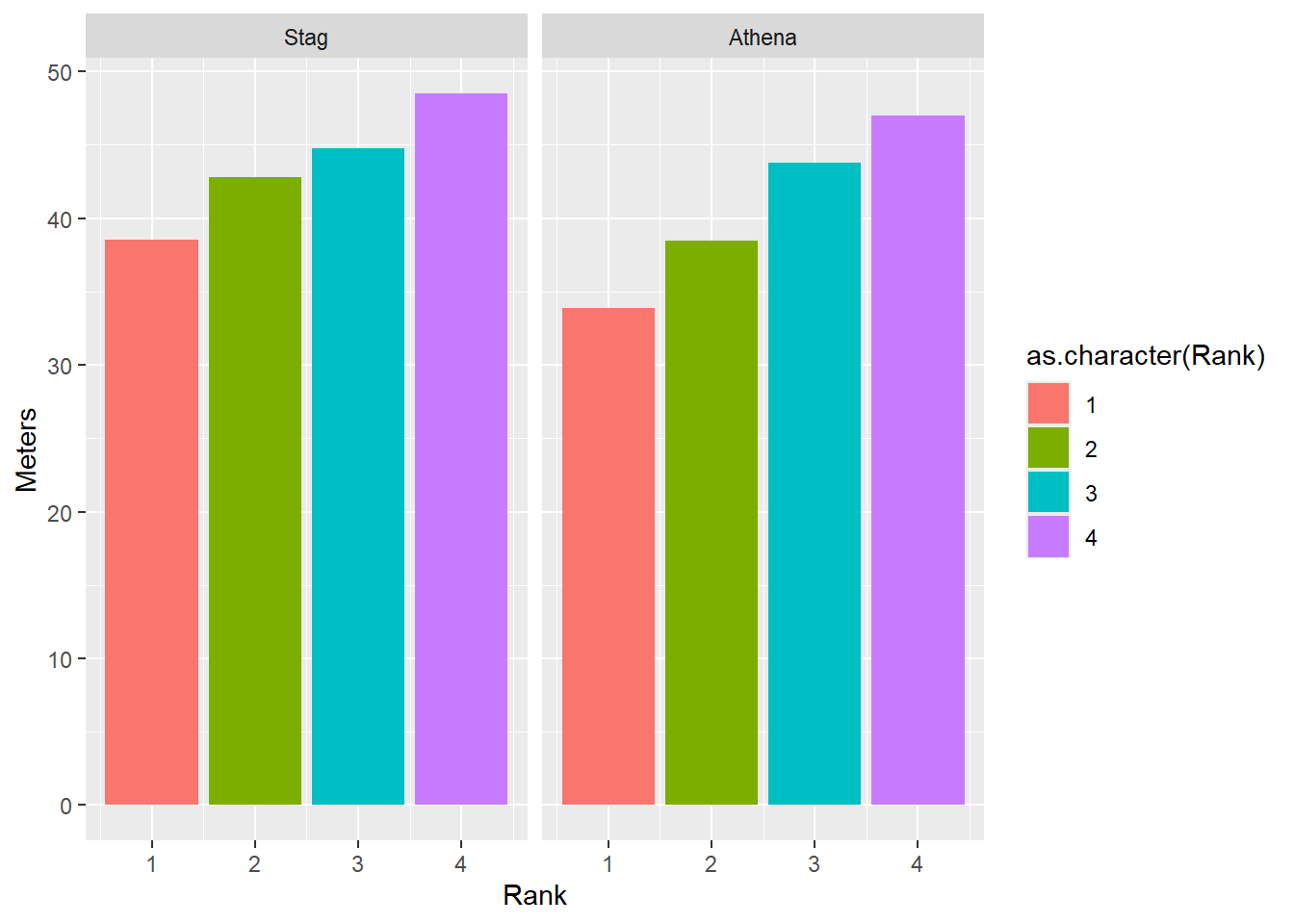
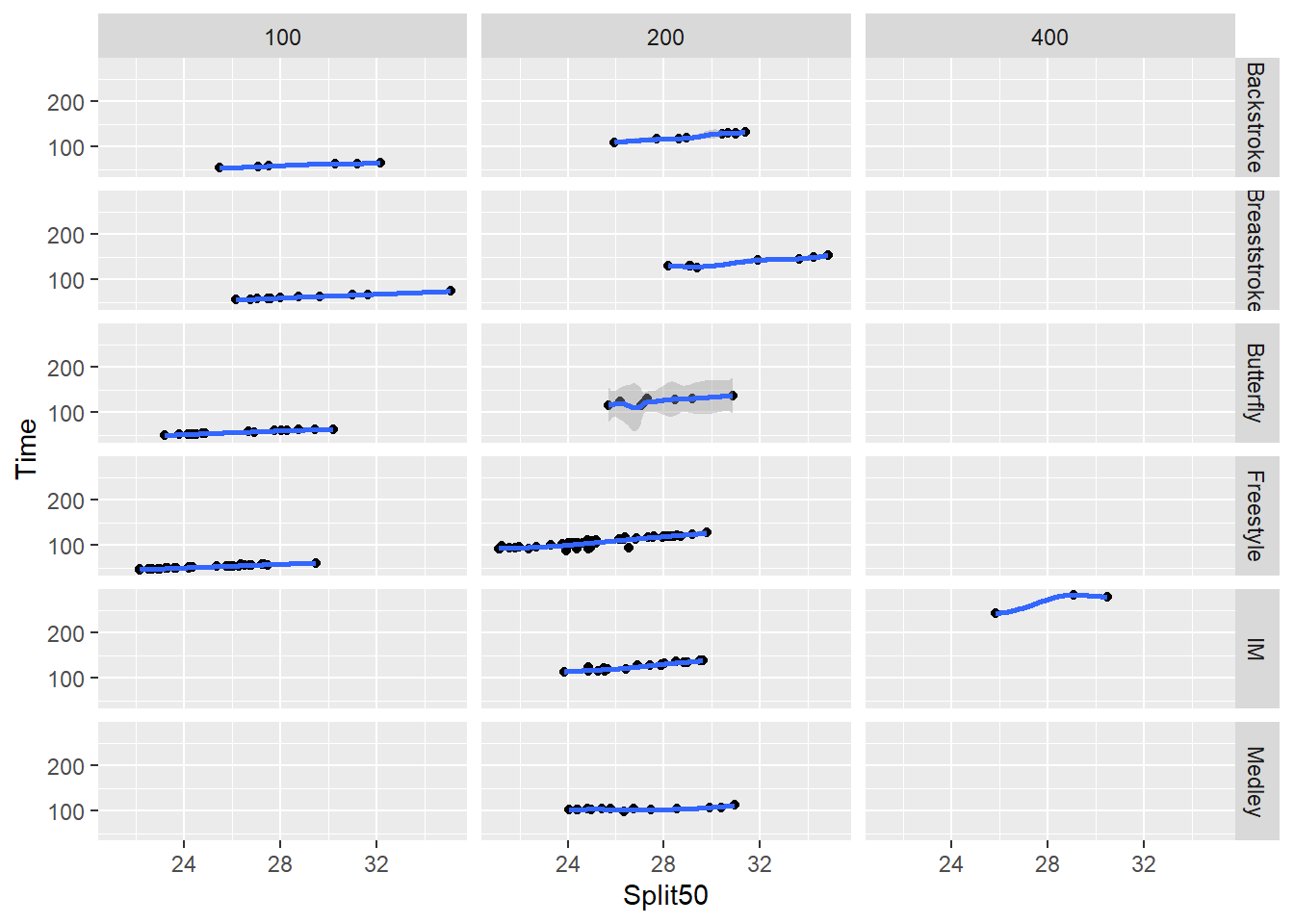
Rows and columns facet_grid(cols = vars())
If your variables allow, you can combine grids with rows and columns.
SWIM <- readr::read_csv("https://github.com/slicesofdata/dataviz24/raw/main/data/swim/cleaned-2023-CMS-Invite.csv")
SWIM |>
filter(Distance > 50 & Distance < 500) |>
ggplot(mapping = aes(x = Split50,
y = Time
)) +
geom_point(position = position_jitter()) +
geom_smooth() +
facet_grid(rows = vars(Event),
cols = vars(Distance)
)
Or by a character vector variable.
SWIM |>
filter(Team != "Mixed") |>
filter(Team != "Freestyle") |>
ggplot(mapping = aes(x = Split50,
y = Time
)) +
geom_point(position = position_jitter()) +
geom_smooth() +
facet_grid(rows = vars(Distance),
cols = vars(Team)
)
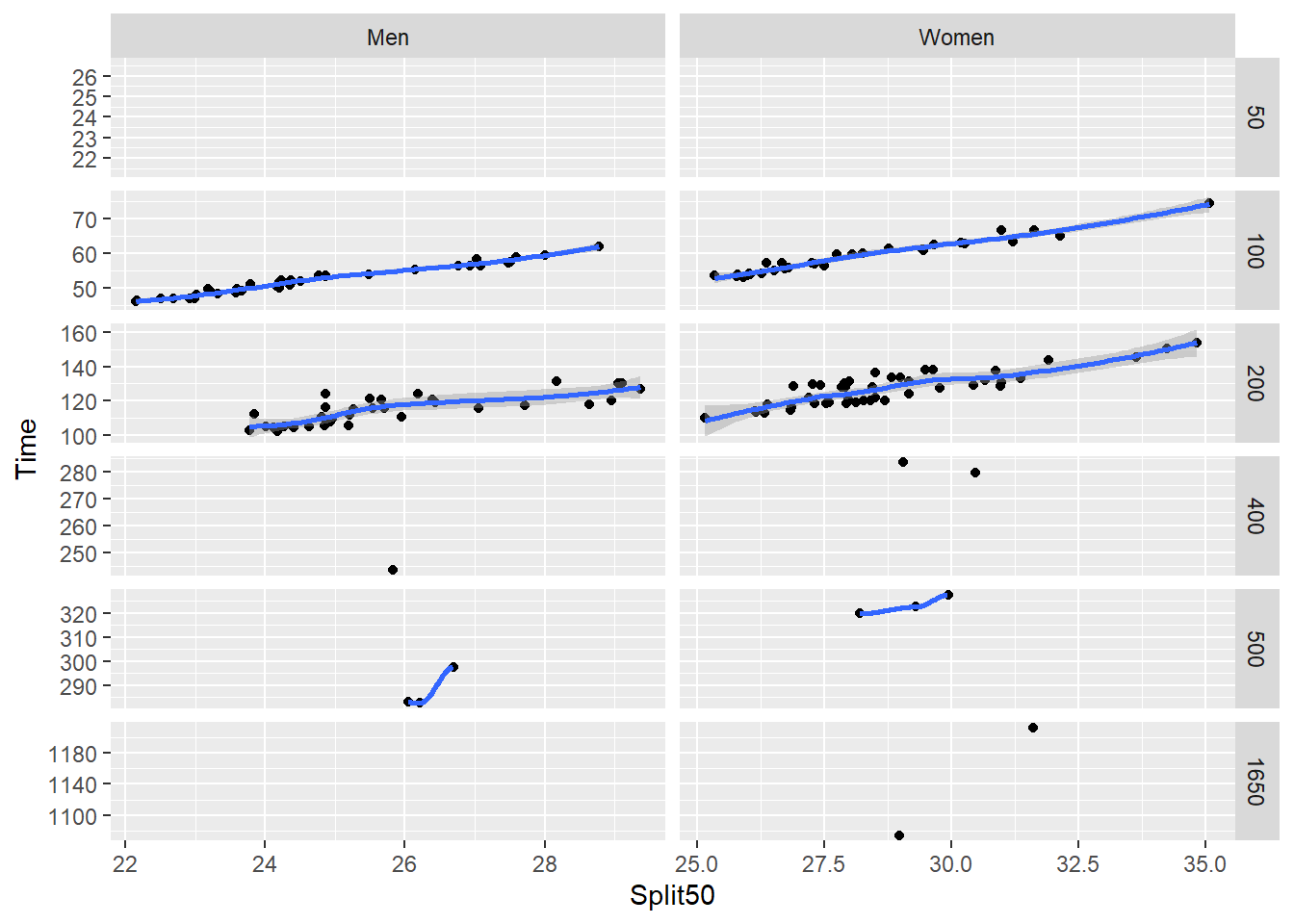
Clearly, we need to clean up the axes for this plot. You can allow scales to vary as was done using facet_wrap() or you can adjust the scales.
SWIM |>
filter(Team != "Mixed") |>
filter(Team != "Freestyle") |>
ggplot(mapping = aes(x = Split50,
y = Time
)) +
geom_point(position = position_jitter()) +
geom_smooth() +
facet_grid(rows = vars(Distance),
cols = vars(Team),
scales = "free"
)
Session Info
sessionInfo()R version 4.4.1 (2024-06-14 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Los_Angeles
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] htmltools_0.5.8.1 DT_0.33 vroom_1.6.5 lubridate_1.9.3
[5] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[9] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1
[13] tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] utf8_1.2.4 generics_0.1.3 lattice_0.22-6 stringi_1.8.4
[5] hms_1.1.3 digest_0.6.36 magrittr_2.0.3 evaluate_0.24.0
[9] grid_4.4.1 timechange_0.3.0 fastmap_1.2.0 Matrix_1.7-0
[13] R.oo_1.26.0 rprojroot_2.0.4 jsonlite_1.8.8 R.utils_2.12.3
[17] mgcv_1.9-1 fansi_1.0.6 scales_1.3.0 cli_3.6.3
[21] rlang_1.1.4 crayon_1.5.3 R.methodsS3_1.8.2 splines_4.4.1
[25] bit64_4.0.5 munsell_0.5.1 withr_3.0.1 yaml_2.3.10
[29] parallel_4.4.1 tools_4.4.1 tzdb_0.4.0 colorspace_2.1-0
[33] pacman_0.5.1 here_1.0.1 curl_5.2.1 vctrs_0.6.5
[37] R6_2.5.1 lifecycle_1.0.4 htmlwidgets_1.6.4 bit_4.0.5
[41] archive_1.1.8 pkgconfig_2.0.3 pillar_1.9.0 gtable_0.3.5
[45] glue_1.7.0 xfun_0.45 tidyselect_1.2.1 rstudioapi_0.16.0
[49] knitr_1.47 farver_2.1.2 nlme_3.1-164 labeling_0.4.3
[53] rmarkdown_2.27 compiler_4.4.1